Abstract
Aims/hypothesis
Oxidative stress is implicated in beta cell glucotoxicity in type 2 diabetes. Inhibitor of differentiation (ID) proteins are transcriptional regulators induced by hyperglycaemia in islets, but the mechanisms involved and their role in beta cells are not clear. Here we investigated whether or not oxidative stress regulates ID levels in beta cells and the role of ID proteins in beta cells during oxidative stress.
Methods
MIN6 cells were cultured in H2O2 or ribose to induce oxidative stress. ID1, ID3 and small MAF proteins (MAFF, MAFG and MAFK) were inhibited using small interfering RNA. Isolated islets from Id1 −/−, Id3 −/− and diabetic db/db mice were used.
Results
ID1–4 expression was upregulated in vivo in the islets of diabetic db/db mice and stimulated in vitro by ribose and H2O2. Id1/3 inhibition reduced the expression of multiple antioxidant genes and potentiated oxidative stress-induced apoptosis. This finding was associated with increased levels of intracellular reactive oxygen species, altered mitochondrial morphology and reduced expression of Tfam, which encodes a mitochondrial transcription factor, and respiratory chain components. Id1/3 inhibition also reduced the expression of small MAF transcription factors (MafF, MafG and MafK), interacting partners of nuclear factor, erythroid 2-like 2 (NFE2L2), master regulator of the antioxidant response. Inhibition of small MAFs reduced the expression of antioxidant genes and potentiated oxidative stress-induced apoptosis, thus recapitulating the effects of Id1/3 inhibition.
Conclusions/interpretation
Our study identifies IDs as a novel family of oxidative stress-responsive proteins in beta cells. IDs are crucial regulators of the adaptive antioxidant–mitochondrial response that promotes beta cell survival during oxidative stress through a novel link to the NFE2L2–small MAF pathway.
Similar content being viewed by others
Introduction
Beta cell failure in type 2 diabetes is characterised by dysfunctional insulin secretion and reduced beta cell mass, which has been linked to an increased rate of apoptosis [1–3]. Several lines of evidence underscore a role for chronic hyperglycaemia—termed ‘glucotoxicity’—in increased beta cell apoptosis. Thus, elevated glucose concentrations trigger apoptosis in cultured islets and beta cell lines [4–8] and in animal models of type 2 diabetes [9–11].
Oxidative stress has been proposed as a central mechanism of hyperglycaemia-induced beta cell demise [12, 13]. Elevated glucose or ribose levels have been shown to increase reactive oxygen species (ROS) production in beta cells [8, 14–16], and islets of diabetic mice exhibit higher ROS content, mitochondrial dysfunction and oxidative damage [15, 17–19].
The vulnerability of beta cells to oxidative stress may be owed to their low expression of several key antioxidant genes, namely Gpx1, Sod1–2 and catalase [20]. However, other genes of the antioxidant arsenal either display strong expression in beta cells (e.g. other Gpx isoforms, Hmox1, Srxn1, Prdxs, G6pdx), or are markedly upregulated under oxidative stress conditions [6, 7, 21, 22]. The signalling pathways that regulate redox status and antioxidant gene expression in beta cells are only partially understood. Nuclear factor, erythroid 2-like 2 (NFE2L2, also known as NRF2) is a primary regulator of the antioxidant response and its activation has been shown to protect beta cells against oxidative damage [23]. NFE2L2 activation of antioxidant gene expression requires its dimerisation with small MAF proteins (MAFF, MAFG and MAFK) [24–26].
The inhibitor of differentiation (ID) proteins are transcriptional regulators that play important roles in both physiology (e.g. development) and pathology (e.g. tumourigenesis). They have been proposed to act as repressors of basic helix-loop-helix transcription factors thereby modulating cell differentiation and proliferation [27, 28]. Previous reports have demonstrated that the expression of ID1 and ID3 is induced by glucose stimulation in human islets and beta cell lines [29, 30]. Further studies link the upregulation of Id1 mRNA levels to hyperglycaemia in islets of diabetic mice [31] and suggest an influence of Id1 expression in insulin secretion [32]. However, the mechanisms underlying the induction of ID proteins by hyperglycaemia and their precise role in beta cell pathophysiology are not clear.
Here, we demonstrate that IDs are novel oxidative stress-responsive proteins in beta cells. We also identify an unexpected role of ID expression in the induction of the antioxidant response under oxidative stress conditions. Inhibition of Id1/3 reduces the expression of multiple antioxidant genes and leads to increased ROS production and apoptosis, and altered mitochondrial function and morphology. Our studies also suggest a mechanism for these effects via a previously unrecognised interaction of IDs with the NFE2L2MAF antioxidant pathway. Our results, therefore, suggest that IDs are key regulators of the adaptive antioxidant–mitochondrial response that promote beta cell survival under oxidative stress.
Methods
Reagents
Ribose was obtained from Sigma (St Louis, MI, USA) and 5-(and-6)-chloromethyl-2′,7′-dichlorodihydrofluorescein diacetate (CM-H2DCFDA) and Deep Red Mitotracker were obtained from Invitrogen (Carlsbad, CA, USA). Control non-targeting and ON-TARGETplus SMARTpool small interfering RNA (siRNA) and transfection reagent DharmaFECT3 were sourced from Thermo Fisher Scientific (Lafayette, CO, USA).
Mice
C57BL/KsJ db/db and their age-matched lean control db/+ mice were taken from the Garvan Institute (Sydney, NSW, Australia) breeding colonies at the age of 14–16 weeks. Wild-type (C57BL/6/129/Sv), Id1 −/− and Id3 −/− mice were bred in-house using animals provided by Robert Benezra (Memorial Sloan-Kettering Cancer Center, New York, NY, USA) and used at the age of 8–10 weeks. All experiments were approved by the Garvan Institute/St Vincent’s Hospital Animal Experimentation Ethics Committee.
Islet culture
Wild-type, Id1 −/− and Id3 −/− islets were cultured in RPMI medium containing 11.1 mmol/l glucose (Invitrogen), 0.2 mmol/l glutamine, 10% heat-inactivated FBS, 100 units/ml penicillin and 100 mg/ml streptomycin in the presence or absence of 50 mmol/l ribose. To assess mitochondrial morphology, islets were dispersed with trypsin and cells were seeded on poly-L-lysine-treated glass coverslips before treatment.
Cell culture and treatment
MIN6 beta cells (P26-39) were grown in Dulbecco’s modified Eagle’s medium (Invitrogen) containing 25 mmol/l glucose, 10 mmol/l HEPES, 10% FCS, 50 units/ml penicillin and 50 mg/ml streptomycin. Cells were transfected with either control, Id1 and/or Id3 or MafF/G/K siRNA. After 24 h, the transfection medium was changed and cells were cultured for 48 h in the absence or presence of H2O2 or ribose.
Real-time RT-PCR
Real-time RT-PCR was performed as previously described [33]. Primer sequences are listed in Electronic Supplementary Material (ESM) Table 1. The value obtained for each specific gene product was normalised to the control gene cyclophilin A and expressed as a fold change of the value in control condition.
Western blots
Western blotting was performed as previously described [32]. Antibodies are listed in ESM Table 2.
Functional assays
Glutathione peroxidase (GPX) activity was determined indirectly as the decrease in NADPH absorption at 340 nm as previously described [34]. Cell death was determined with the use of a Cell Death Detection ELISA (Roche Diagnostics, Castle Hill, NSW, Australia) [6]. ATP generation was determined using ATPlite kit (PerkinElmer, Melbourne, VIC, Australia). Oxygen consumption was measured using a Clark-type oxygen electrode system as previously described [34]. For ROS measurement, cells were incubated with 2 μmol/l CM-H2DCFDA or DMSO for 30 min at 37°C. Fluorescence intensity was measured (ex/em at 485/520 nm) using a plate reader (FluoStar OPTIMA, BMG Lab Technologies, Mornington, VIC, Australia).
Immunofluorescence
Immunostaining was performed as previously described [35]. Images were acquired using a Leica SP8 confocal microscope.
Statistical analysis
Results are given as mean ± SEM for the indicated number of experiments. Statistical significance was assessed by unpaired two-tailed Student’s t test, one-way ANOVA and a post-test of Newman–Keuls or two-way ANOVA and a post-test of Bonferroni.
Results
ID expression is increased in the islets of diabetic mice in parallel with the antioxidant response
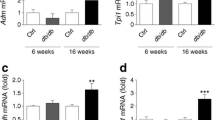
By immunofluorescence on histological pancreatic sections we observed a marked increase in the expression of ID1 and ID3 in the islets of diabetic db/db mice in comparison with normoglycaemic control db/+ mice (Fig. 1a,b). These effects were paralleled by increased expression of NFE2L2, a master transcriptional regulator of antioxidant gene expression (Fig. 1c). In agreement with these observations, mRNA levels of Id1–4 were upregulated in the islets of db/db mice (Fig. 1d–g). Furthermore, mRNA levels of the antioxidant genes Hmox1, Gpx1–2, Txnrd1, G6pdx, Srxn1 and Prdx1–4 were increased (Fig. 1h–q). The mRNA levels of Nfe2l2 were unchanged (Fig. 1r). These results demonstrate an in vivo association between the upregulation of IDs and the antioxidant response in islets of diabetic db/db mice.
ID protein expression is upregulated in the islets of diabetic mice in parallel with antioxidant genes. Immunostaining of (a) ID1, (b) ID3 and (c) NFE2L2 in islets of control db/+ and diabetic db/db mice. Images are representative of four animals per group. (d–r) Changes in the mRNA levels of Id1–4 and antioxidant genes in the islets of db/+ and db/db mice. n = 6–10 animals per group. *p < 0.05, **p < 0.01, ***p < 0.001 vs db/+. Scale bar, 50 μm
Id genes are induced by oxidative stress in beta cells
MIN6 cells [32] were treated with the pro-oxidant agents H2O2 or ribose. Ribose is a sugar that produces ROS more potently and in a shorter time than glucose and is an established model in which to study beta cell glucotoxicity in vitro [14, 36]. Treatment with H2O2 (Fig. 2a–e) and ribose (Fig. 2f–j) increased Id1–4 and Hmox1 mRNA levels in a concentration-dependent manner. Accordingly, ribose treatment increased the (peri)nuclear expression of ID1 (Fig. 2k) and ID3 proteins (Fig. 2l).
Oxidative stress upregulates Id genes in beta cells. MIN6 cells were cultured for 48 h in the absence or presence of (a–e) H2O2 (100–300 μmol/l) or (f–j) ribose (5–50 mmol/l). n = 3 experiments. *p < 0.05, **p < 0.01, ***p < 0.001 vs control. Immunostaining of (k) ID1 (green) and (l) ID3 (red). Images are representative of four experiments. Scale bar, 10 μm
Id1 and Id3 mRNA levels were induced by ribose in a time-dependent manner. The effect of ribose was significant at 6 h and reached a maximum at 48 h (Fig. 3a,b). The mRNA levels of antioxidant genes were also induced by ribose in a time-dependent manner. Hmox1, Srxn1 and, to a lesser extent, Gpx2 were induced at 6 h and reached the maximum at 24 h (Fig. 3c–e) while Prdx1 was significantly induced between 24–48 h and reached the maximum at 48 h (Fig. 3f). In parallel, Nfe2l2 tended to be induced at 6 h and reached the maximum at 48 h (Fig. 3g). The mRNA levels of the proapoptotic gene Bax were upregulated by ribose treatment slightly later than Id and antioxidant genes (Fig. 3h). These findings suggest a close association between IDs and antioxidant gene expression under oxidative stress conditions in MIN6 beta cells.
Oxidative stress time-dependently increases mRNA levels of Id1, Id3, antioxidant and proapoptotic genes. MIN6 cells were cultured from 0–48 h in the absence (open circles) or presence (closed circles) of 50 mmol/l ribose. n = 3 experiments. *p < 0.05, **p < 0.01, ***p < 0.001 vs time 0; † p < 0.05, †† p < 0.01, ††† p < 0.001 vs same group in the absence of ribose
Furthermore, the ribose-induced upregulation of mRNA levels for Id genes was prevented by treatment with the antioxidant MnTBAP (ESM Fig. 1a–j). Together these results demonstrate that IDs are novel oxidative stress-responsive proteins in MIN6 beta cells.
Inhibition of Id1 and/or Id3 leads to global attenuation of the antioxidant response
To explore the role of ID induction under oxidative stress in beta cells, we used siRNA to inhibit the expression of Id1 and/or Id3 (Fig. 4a,b). Interestingly, in ribose-treated cells, inhibition of Id1 significantly reduced the mRNA levels of Hmox1. When both Id1 and Id3 were inhibited a stronger effect was observed (Fig. 4c). This additive inhibitory effect was observed for all the other antioxidant genes studied, including Gpx1–2, Srxn1, Prdx1–4, Txnrd1 and G6pdx (Fig. 4d–l), while Nfe2l2 was not significantly affected (Fig. 4m). In agreement with this finding, Id1 inhibition decreased H2O2- and ribose-induced upregulation of heme oxygenase 1 (HMOX1) protein levels and this reduction was stronger after inhibition of both isoforms (Fig. 4n). We also found that inhibition of Id1/3 reduced the ribose-mediated upregulation of superoxide dismutase 2 (SOD2) protein levels. Moreover, ribose-induced upregulation of GPX activity was partially prevented by approximately 21% after Id1/3 inhibition (Fig. 4o). These results indicate a striking association between Id1/3 inhibition and a global attenuation of the antioxidant response.
Inhibition of Id1 and/or Id3 attenuates the global antioxidant response. MIN6 cells transfected with either control siRNA (si-C), siRNA against Id1 (si-Id1), Id3 (si-Id3) or both (si-Id1/3) were cultured for 48 h in the absence (white bars) or presence of 50 mmol/l ribose (black bars). (a–m) Changes in mRNA levels. Graphs are representative of three experiments. *p < 0.05, **p < 0.01, ***p < 0.001 vs same group in the absence of ribose; † p < 0.05, †† p < 0.01, ††† p < 0.001 vs control siRNA. (n) Changes in protein levels. n = 3–5 experiments. C, control; H, H2O2 100 μmol/l; R, ribose 50 mmol/l. (o) Changes in GPX enzymatic activity. n = 5 experiments. *p < 0.05, ***p < 0.001 vs same group in the absence of ribose. U, untreated
Inhibition of Id1 and/or Id3 increases ROS levels, apoptosis and oxidative damage
We assessed the influence of Id inhibition on ROS generation and cell viability. The attenuated antioxidant response may occur as a consequence of reduced ROS levels or it may lead to increased ROS generation. Interestingly, Id1/3 inhibition strongly increased basal and ribose-stimulated H2O2 levels by approximately 2.4- and 1.8-fold respectively (Fig. 5a). In line with these results, Id1/3 inhibition doubled basal and H2O2- and ribose-induced apoptosis (Fig. 5b). Moreover, in cultured islets basal and ribose-induced apoptosis tended to be potentiated in Id1 −/− islets and ribose-induced apoptosis was significantly increased in Id3 −/− islets (Fig. 5c). The inhibition of Id1 or Id3 alone in MIN6 beta cells also potentiated ribose-induced apoptosis but the strongest effect was observed when both isoforms were inhibited (ESM Fig. 2). The absence of Id1/3 was also associated with increased mRNA levels of Bax and DNA-damage inducible transcript 2 (Fig. 5d,e), and reduced mRNA levels of the anti-apoptotic genes Bcl2 and Bcl-xl (Fig. 5f,g). Accordingly, DNA-damage was increased after inhibition of Id1/3. Thus, 8-hydroxyguanosine (8OHdG) immunostaining was barely detected in untreated cells and markedly increased by ribose treatment (Fig. 5h,i). This staining was further increased upon Id1/3 inhibition in control and ribose-treated cells (Fig. 5j,k). Altogether, these results demonstrate that the inhibition of Id1/3 leads to increased ROS levels, oxidative damage and apoptosis.
Inhibition of Id1 and Id3 increases ROS levels, oxidative damage and apoptosis. MIN6 cells transfected with either control siRNA (si-C) or siRNA against Id1 and Id3 (si-Id1/3) were cultured for 48 h in the absence (white bars) or presence of 50 mmol/l ribose (black bars) or 100 μmol/l H2O2 (grey bars). Changes in (a) intracellular ROS and (b) apoptosis levels. (c) Changes in apoptosis levels in isolated wild-type (WT), Id1 −/− and Id3 −/− mouse islets cultured for 48 h in the absence (white bars) or presence of 50 mmol/l ribose (black bars). (d–g) Changes in mRNA levels in MIN6 cells. n = 3–4 experiments. *p < 0.05, **p < 0.01, ***p < 0.001 vs same group in the absence of ribose or H2O2; † p < 0.05, †† p < 0.01, ††† p < 0.001 vs control siRNA or wild-type. (h–k) Immunostaining of 8OHdG (red). Images are representative of three experiments. Scale bar, 10 μm
Inhibition of Id1 and/or Id3 impairs mitochondrial homeostasis
An association between increased ROS generation and altered mitochondrial function has been demonstrated in islets of diabetic animals [15, 19]. We assessed mitochondrial function by measuring oxygen consumption and ATP generation. Ribose treatment tended to reduce oxygen consumption. After inhibition of Id1/3, oxygen consumption tended to be lower in untreated cells and was significantly reduced by approximately 20% after ribose treatment (Fig. 6a). Ribose treatment also markedly reduced ATP levels but this effect was stronger after inhibition of Id1/3 (Fig. 6b).
Inhibition of Id1 and Id3 impairs mitochondrial homeostasis and alters mitochondrial morphology. MIN6 cells transfected with either control siRNA (si-C) or siRNA against Id1 and Id3 (si-Id1/3) were cultured for 48 h in the absence (white bars) or presence of 50 mmol/l ribose (black bars) or 100 μmol/l H2O2 (grey bars). (a) Changes in oxygen consumption, (b) ATP levels, (c–h) protein levels of ETC complex components and (i) mRNA levels of the mitochondrial transcription factor Tfam. n = 3–5 experiments. *p < 0.05, **p < 0.01, ***p < 0.001 vs same group in the absence of ribose; † p < 0.05, †† p < 0.01, ††† p < 0.001 vs control siRNA. C, control; H, H2O2 100 μmol/l; R, ribose 50 mmol/l. (j–m) Mitochondrial (Mito) morphology (red) in MIN6 cells cultured for 48 h in the absence or presence of 50 mmol/l ribose. Images are representative of five experiments. Scale bar, 10 μm
This alteration in mitochondrial function was associated with changes in the expression of electron transport chain (ETC) components. Thus, inhibition of Id1/3 tended to decrease the expression of CI (subunit NDUFB8), CII (subunit SDHB) and CIII (subunit UQCRC2), significantly decreased the expression of CV (subunit ATP5A), and strongly decreased the expression of CIV (subunit MTCO1) (Fig. 6c–h). On the other hand, the expression of Ucp2 was not affected by Id1/3 inhibition (ESM Fig. 3a). MTCO1, a catalytic subunit of cytochrome c oxidase, is regulated by transcription factor A, mitochondrial (TFAM). The mRNA levels of Tfam were upregulated by ribose treatment (Fig. 6i) as well as in the islets of diabetic db/db mice (ESM Fig. 3b). Interestingly, inhibition of Id1/3 reduced the mRNA levels of Tfam (Fig. 6i).
Increased ROS production has been associated with altered mitochondrial morphology [15, 19]. Exposure to ribose modified the mitochondrial morphology into a more condensed-globular pattern (Fig. 6j,k). Interestingly, Id1/3 inhibition altered mitochondrial morphology both in untreated cells (Fig. 6l) and after ribose treatment (Fig. 6m). Similarly, in primary beta cells from wild-type mice, mitochondrial morphology was altered by ribose treatment (Fig. 7a). In beta cells from Id3 −/− mice, altered morphology was observed under control and ribose conditions (Fig. 7b).
Together, these results show that the inhibition of Id1/3 is associated with altered mitochondrial function and morphology. These alterations were associated with reduced expression of components of the ETC, particularly MTCO1, as well as the upstream transcription factor Tfam.
Inhibition of Id1 and Id3 reduces the expression of MAFs, interacting partners of NFE2L2
Under control conditions, NFE2L2 was barely detected in the cytosol while ribose stimulation increased its nuclear localisation (Fig. 8a,b). Surprisingly, inhibition of Id1/3 increased NFE2L2 nuclear expression under control conditions and after ribose treatment (Fig. 8c,d) without affecting the expression of the cytoplasmic adaptor protein kelch-like ECH-associated protein (ESM Fig. 4a,b). This observation is in agreement with the finding of increased oxidative stress in the absence of Id1/3, but does not explain the inhibitory effect on antioxidant gene expression. We therefore hypothesised that IDs may affect an interacting partner of NFE2L2 (Fig. 8e). Atf4 [37] and c-Jun [38] were potential candidates. However, Atf4 mRNA levels (ESM Fig. 5a) and phospho-c-JUN protein levels (ESM Fig. 5b) were unaffected by Id1/3 inhibition. The other potential candidates were the small MAF proteins [24–26, 38]. We found that ribose treatment markedly increased MafF and MafK mRNA levels. Interestingly, Id1/3 inhibition strongly prevented this increase (Fig. 8f,g). MafG mRNA levels were slightly increased by ribose treatment and slightly reduced by Id1/3 inhibition (Fig. 8h). Furthermore, MAFK protein levels were reduced by Id1/3 inhibition (Fig. 8i). These findings demonstrate that the inhibition of Id1/3 leads to reduced expression of small MAF proteins. Confirming the potential physiological relevance of these findings, MafF and MafK mRNA levels were markedly increased in the islets of diabetic db/db mice while the mRNA levels of MafG were not significantly affected (Fig. 8j–l). These findings suggest a novel mechanism whereby IDs regulate antioxidant gene expression by modifying the expression of small MAF proteins, NFE2L2-interacting partners.
Inhibition of Id1 and Id3 increases the nuclear accumulation of NFE2L2 while reducing the expression of its interacting partners, the MAFs. MIN6 cells transfected with either control siRNA (si-C) or siRNA against Id1 and Id3 (si-Id1/3) were cultured for 48 h in the absence (white bars) or presence (black bars) of 50 mmol/l ribose. (a–d) Immunostaining of NFE2L2. Images are representative of three experiments. Scale bar, 10 μm. (e) Analysis of NFE2L2-interacting partners using Cytoscape (2.8.3) and MiMl plug-in (www.cytoscape.org). (f–h) Changes in MafF, MafK and MafG mRNA levels. n = 4 experiments. *p < 0.05, ***p < 0.001 vs same group in the absence of ribose; †† p < 0.01, ††† p < 0.001 vs control siRNA. (i) Changes in MAFK protein levels. n = 4 experiments. C, control; R, ribose 50 mmol/l. (j–l) Changes in mRNA levels in islets of db/+ and db/db mice. n = 6 animals/group. *p < 0.05, ***p < 0.001 vs db/+
Inhibition of MAFs reduces antioxidant gene expression and potentiates apoptosis, recapitulating the effects of Id1/3 inhibition
To verify whether the inhibition of small MAF proteins recapitulate the effects of Id1/3 inhibition on antioxidant gene expression, we used siRNA against MafF, MafK and MafG. By this means, the ribose-mediated upregulation of MafF, MafK and MafG was inhibited to a similar extent to that observed after inhibition of Id1/3 (Fig. 9a–c). Strikingly, the inhibition of MAFs resulted in reduced mRNA levels of several antioxidant genes, including Hmox1, Gpx1, Txnrd1, Prdx2–3 and, to a lesser degree, G6pdx (Fig. 9d–i). Furthermore, inhibition of MAFs potentiated ribose-induced apoptosis by approximately 1.8-fold (Fig. 9j).
Inhibition of MAFs reproduces the effects of Id1/3 inhibition on antioxidant gene expression and apoptosis. MIN6 cells transfected with either control siRNA (si-C) or siRNA against MafF, MafK and MafG (si-FGK) were cultured for 48 h in the absence (white bars) or presence of 50 mmol/l ribose (black bars). Changes in (a–i) mRNA levels and (j) apoptosis. n = 3–4 experiments. **p < 0.01, ***p < 0.001 vs same group in the absence of ribose; † p < 0.05, †† p < 0.01, ††† p < 0.001 vs control siRNA. (k) The proposed model: IDs are oxidative stress-responsive proteins that regulate redox status, mitochondrial integrity and beta cell survival. IDs maintain the adaptive mitochondrial–antioxidant response during oxidative stress via (1) upregulation of TFAM and components of the ETC; and (2) upregulation of NFE2L2-interacting partners small MAF proteins and subsequent induction of antioxidant genes
These results demonstrate for the first time the importance of small MAF proteins (F/K/G) in the regulation of antioxidant gene expression and beta cell survival under conditions of oxidative stress. Our results strongly suggest that Id1/3 regulate antioxidant gene expression via the regulation of Maf expression, thereby providing a novel mechanism of the cellular antioxidant response.
Discussion
We have unveiled a novel cellular role for IDs in the regulation of redox status, mitochondrial integrity and beta cell survival under oxidative stress conditions. We have shown for the first time that IDs are oxidative stress-responsive genes in beta cells. All four members of the ID family were upregulated in the islets of diabetic db/db mice. This induction was paralleled by a global upregulation of the antioxidant response. Inhibition of Id1/3 in beta cells led to mitochondrial impairment and global attenuation of the antioxidant response. Since oxidative stress is a major downstream mechanism of beta cell glucotoxicity [13], our results provide important insight into the molecular mechanisms regulating beta cell survival in type 2 diabetes.
IDs and the antioxidant response
NFE2L2 is a key regulator of antioxidant gene expression and cellular homeostasis [39]. Unexpectedly, we found that the inhibition of Id1/3 markedly increased NFE2L2 expression and nuclear localisation. This interesting observation, which is in line with the increased ROS accumulation under these conditions, suggested that Id1/3 may affect an interacting partner rather than NFE2L2 itself. Indeed, we found that inhibition of Id1/3 inhibited the expression of small MAF proteins F, K and G, known interacting partners of NFE2L2 that play an important role in the regulation of antioxidant gene expression [24–26, 40]. Interestingly, inhibition of MafF/G/K reduced the expression of several antioxidant genes and potentiated ribose-induced apoptosis, thereby mimicking the effects of Id1/3 inhibition.
These striking findings provide a novel regulatory mechanism of the antioxidant response. Our results strongly suggest that Id1/3 regulate antioxidant gene expression through the regulation of small MAF protein expression. Our results highlight the important role of small MAF proteins in beta cells. While recent reports have focused on the role of NFE2L2 in diabetes and beta cell pathophysiology [23, 41], less attention has been given to the role of small MAF proteins. Yet, our results show that increased expression and nuclear accumulation of NFE2L2 alone, in the absence of MAFs, was insufficient to maintain adequate expression of antioxidant genes under oxidative stress.
IDs and mitochondria
Mitochondria have been shown to be a primary target for oxidative damage. Thus, acute exposure of rat islets and INS1 cells to H2O2 has been shown to impair mitochondrial function, reduce Tfam and ETC component expression, especially MTCO1, and increase mitochondrial ROS production and apoptosis [42]. These observations mirror the effects of Id1/3 inhibition. The global attenuation of antioxidant gene expression induced by Id1/3 inhibition may lead to ROS accumulation and mitochondrial alterations that enhance ROS generation, thus further exacerbating oxidative stress/damage and apoptosis. Alternatively, the downregulation of Tfam and ETC components by Id1/3 inhibition may slow electron flow leading to enhanced ROS generation. Indeed, it is well established that beta cells generate high ROS levels when cultured in the presence of a low non-stimulatory glucose concentration in association with increased expression of antioxidant genes and apoptosis [6, 43, 44]. The resulting oxidative stress is further aggravated by the global attenuation of the antioxidant response. This view is supported by the observation that Id1/3 inhibition leads to enhanced ROS formation, increased oxidative damage and potentiation of apoptosis under basal conditions in the absence of any ribose or H2O2 stimulation. Moreover, in our model, chronic stimulation with H2O2 or ribose increases TFAM and ETC component expression. In line with this observation, mRNA levels of Tfam were upregulated in the islets of diabetic mice despite the presence of oxidative stress. Either way, these alterations were associated with altered mitochondrial morphology. The latter has been observed in beta cells of type 2 diabetes both in animal models and in humans, as well as in in vitro models of nutrient oversupply and oxidative stress [15, 19, 42, 45–47].
Potential exploitation of these new findings to protect beta cells
Although antioxidant supplementation has been shown to be promising in type 2 diabetes animal models, it was overall ineffective in humans. Consequently, the activation of endogenous antioxidant genes could be a more promising strategy to reduce ROS-mediated apoptosis in beta cells. The identification of IDs as key upstream regulators of the antioxidant response, besides their parallel role in mitochondria, may serve for the development of a novel therapeutic strategy to protect beta cells against oxidative stress. IDs are known downstream targets of the bone morphogenetic protein (BMP) signalling pathway. Emerging evidence indicates an important role of this pathway in beta cells [48–50]. However, one should keep in mind that the activation of IDs may also interfere with beta cell differentiation. We have previously shown that Id1 was induced by elevated lipids in beta cells and plays a role in lipid-induced impairment of insulin secretion and differentiation [32]. Moderate physiological stimulation of this pathway may exert beneficial effects under oxidative stress while avoiding the alteration of the beta cell phenotype. Interestingly, our other studies show that low concentrations of recombinant BMP stimulated the expression of Id1/3 in parallel with a significant protection against ribose-induced apoptosis in MIN6 cells (M. Bensellam, D. R. Laybutt, unpublished results).
In conclusion, we have identified IDs as a novel family of oxidative stress-responsive proteins in beta cells. We have demonstrated that Id1/3 are crucial for the maintenance of an adaptive mitochondrial-antioxidant response that promotes beta cell survival under oxidative stress via a novel link to the NFE2L2–MAF signalling pathway. These novel findings may help the development of therapeutic strategies to protect beta cells against oxidative stress.
Abbreviations
- 8OHdG:
-
8-Hydroxyguanosine
- BMP:
-
Bone morphogenetic protein
- ETC:
-
Electron transport chain
- GPX:
-
Glutathione peroxidase
- HMOX1:
-
Heme oxygenase 1
- ID:
-
Inhibitor of differentiation
- NFE2L2:
-
Nuclear factor, erythroid 2-like 2
- ROS:
-
Reactive oxygen species
- siRNA:
-
Small interfering RNA
- SOD2:
-
Superoxide dismutase 2
- TFAM:
-
Transcription factor A, mitochondrial
References
Sakuraba H, Mizukami H, Yagihashi N, Wada R, Hanyu C, Yagihashi S (2002) Reduced beta-cell mass and expression of oxidative stress-related DNA damage in the islet of Japanese type II diabetic patients. Diabetologia 45:85–96
Rahier J, Guiot Y, Goebbels RM, Sempoux C, Henquin JC (2008) Pancreatic β-cell mass in European subjects with type 2 diabetes. Diabetes Obes Metab 10(Suppl 4):32–42
Butler AE, Janson J, Bonner-Weir S, Ritzel R, Rizza RA, Butler PC (2003) β-cell deficit and increased β-cell apoptosis in humans with type 2 diabetes. Diabetes 52:102–110
Maedler K, Spinas GA, Lehmann R et al (2001) Glucose induces β-cell apoptosis via upregulation of the Fas receptor in human islets. Diabetes 50:1683–1690
DelGuerra S, Grupillo M, Masini M et al (2007) Gliclazide protects human islet beta-cells from apoptosis induced by intermittent high glucose. Diabetes Metab Res Rev 23:234–238
Bensellam M, van Lommel L, Overbergh L, Schuit FC, Jonas JC (2009) Cluster analysis of rat pancreatic islet gene mRNA levels after culture in low-, intermediate- and high-glucose concentrations. Diabetologia 52:463–476
Jonas JC, Bensellam M, Duprez J, Elouil H, Guiot Y, Pascal SM (2009) Glucose regulation of islet stress responses and β-cell failure in type 2 diabetes. Diabetes Obes Metab 11(Suppl 4):65–81
Wali JA, Rondas D, McKenzie MD et al (2014) The proapoptotic BH3-only proteins Bim and Puma are downstream of endoplasmic reticulum and mitochondrial oxidative stress in pancreatic islets in response to glucotoxicity. Cell Death Dis 5:e1124
Donath MY, Gross DJ, Cerasi E, Kaiser N (1999) Hyperglycemia-induced β-cell apoptosis in pancreatic islets of Psammomys obesus during development of diabetes. Diabetes 48:738–744
Song B, Scheuner D, Ron D, Pennathur S, Kaufman RJ (2008) Chop deletion reduces oxidative stress, improves β cell function, and promotes cell survival in multiple mouse models of diabetes. J Clin Invest 118:3378–3389
Kluth O, Mirhashemi F, Scherneck S et al (2011) Dissociation of lipotoxicity and glucotoxicity in a mouse model of obesity associated diabetes: role of forkhead box O1 (FOXO1) in glucose-induced beta cell failure. Diabetologia 54:605–616
Robertson RP, Harmon JS (2006) Diabetes, glucose toxicity, and oxidative stress: a case of double jeopardy for the pancreatic islet β cell. Free Radic Biol Med 41:177–184
Bensellam M, Laybutt DR, Jonas JC (2012) The molecular mechanisms of pancreatic beta-cell glucotoxicity: recent findings and future research directions. Mol Cell Endocrinol 364:1–27
Tanaka Y, Tran PO, Harmon J, Robertson RP (2002) A role for glutathione peroxidase in protecting pancreatic β cells against oxidative stress in a model of glucose toxicity. Proc Natl Acad Sci U S A 99:12363–12368
Bindokas VP, Kuznetsov A, Sreenan S, Polonsky KS, Roe MW, Philipson LH (2003) Visualizing superoxide production in normal and diabetic rat islets of Langerhans. J Biol Chem 278:9796–9801
Sakai K, Matsumoto K, Nishikawa T et al (2003) Mitochondrial reactive oxygen species reduce insulin secretion by pancreatic β-cells. Biochem Biophys Res Commun 300:216–222
Ihara Y, Toyokuni S, Uchida K et al (1999) Hyperglycemia causes oxidative stress in pancreatic β-cells of GK rats, a model of type 2 diabetes. Diabetes 48:927–932
Gorogawa S, Kajimoto Y, Umayahara Y et al (2002) Probucol preserves pancreatic β-cell function through reduction of oxidative stress in type 2 diabetes. Diabetes Res Clin Pract 57:1–10
Lu H, Koshkin V, Allister EM, Gyulkhandanyan AV, Wheeler MB (2010) Molecular and metabolic evidence for mitochondrial defects associated with β-cell dysfunction in a mouse model of type 2 diabetes. Diabetes 59:448–459
Lenzen S, Drinkgern J, Tiedge M (1996) Low antioxidant enzyme gene expression in pancreatic islets compared with various other mouse tissues. Free Radic Biol Med 20:463–466
Laybutt DR, Kaneto H, Hasenkamp W et al (2002) Increased expression of antioxidant and antiapoptotic genes in islets that may contribute to β-cell survival during chronic hyperglycemia. Diabetes 51:413–423
Lacraz G, Figeac F, Movassat J et al (2009) Diabetic β-cells can achieve self-protection against oxidative stress through an adaptive up-regulation of their antioxidant defenses. PLoS ONE 4:e6500
Yagishita Y, Fukutomi T, Sugawara A et al (2014) Nrf2 protects pancreatic beta-cells from oxidative and nitrosative stress in diabetic model mice. Diabetes 63:605–618
Katsuoka F, Motohashi H, Ishii T, Aburatani H, Engel JD, Yamamoto M (2005) Genetic evidence that small maf proteins are essential for the activation of antioxidant response element-dependent genes. Mol Cell Biol 25:8044–8051
Hirotsu Y, Katsuoka F, Funayama R et al (2012) Nrf2-MafG heterodimers contribute globally to antioxidant and metabolic networks. Nucleic Acids Res 40:10228–10239
Tanigawa S, Lee CH, Lin CS et al (2013) Jun dimerization protein 2 is a critical component of the Nrf2/MafK complex regulating the response to ROS homeostasis. Cell Death Dis 4:e921
Ruzinova MB, Benezra R (2003) Id proteins in development, cell cycle and cancer. Trends Cell Biol 13:410–418
Nair R, Teo WS, Mittal V, Swarbrick A (2014) ID proteins regulate diverse aspects of cancer progression and provide novel therapeutic opportunities. Mol Ther 22:1407–1415
Webb GC, Akbar MS, Zhao C, Steiner DF (2000) Expression profiling of pancreatic β cells: glucose regulation of secretory and metabolic pathway genes. Proc Natl Acad Sci U S A 97:5773–5778
Wice BM, Bernal-Mizrachi E, Permutt MA (2001) Glucose and other insulin secretagogues induce, rather than inhibit, expression of Id-1 and Id-3 in pancreatic islet beta cells. Diabetologia 44:453–463
Kjorholt C, Akerfeldt MC, Biden TJ, Laybutt DR (2005) Chronic hyperglycemia, independent of plasma lipid levels, is sufficient for the loss of β-cell differentiation and secretory function in the db/db mouse model of diabetes. Diabetes 54:2755–2763
Akerfeldt MC, Laybutt DR (2011) Inhibition of Id1 augments insulin secretion and protects against high-fat diet-induced glucose intolerance. Diabetes 60:2506–2514
Chan JY, Luzuriaga J, Bensellam M, Biden TJ, Laybutt DR (2013) Failure of the adaptive unfolded protein response in islets of obese mice is linked with abnormalities in beta-cell gene expression and progression to diabetes. Diabetes 62:1557–1568
Montgomery MK, Osborne B, Brown SH et al (2013) Contrasting metabolic effects of medium- versus long-chain fatty acids in skeletal muscle. J Lipid Res 54:3322–3333
Bensellam M, Duvillie B, Rybachuk G et al (2012) Glucose-induced O2 consumption activates hypoxia inducible factors 1 and 2 in rat insulin-secreting pancreatic beta-cells. PLoS ONE 7:e29807
Kaneto H, Fujii J, Myint T et al (1996) Reducing sugars trigger oxidative modification and apoptosis in pancreatic β-cells by provoking oxidative stress through the glycation reaction. Biochem J 320(Pt 3):855–863
He CH, Gong P, Hu B et al (2001) Identification of activating transcription factor 4 (ATF4) as an Nrf2-interacting protein. Implication for heme oxygenase-1 gene regulation. J Biol Chem 276:20858–20865
Newman JR, Keating AE (2003) Comprehensive identification of human bZIP interactions with coiled-coil arrays. Science 300:2097–2101
Itoh K, Tong KI, Yamamoto M (2004) Molecular mechanism activating Nrf2-Keap1 pathway in regulation of adaptive response to electrophiles. Free Radic Biol Med 36:1208–1213
Itoh K, Chiba T, Takahashi S et al (1997) An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem Biophys Res Commun 236:313–322
Uruno A, Furusawa Y, Yagishita Y et al (2013) The Keap1-Nrf2 system prevents onset of diabetes mellitus. Mol Cell Biol 33:2996–3010
Li N, Brun T, Cnop M, Cunha DA, Eizirik DL, Maechler P (2009) Transient oxidative stress damages mitochondrial machinery inducing persistent beta-cell dysfunction. J Biol Chem 284:23602–23612
Martens GA, Cai Y, Hinke S, Stange G, van de Casteele M, Pipeleers D (2005) Glucose suppresses superoxide generation in metabolically responsive pancreatic beta cells. J Biol Chem 280:20389–20396
Roma LP, Pascal SM, Duprez J, Jonas JC (2012) Mitochondrial oxidative stress contributes differently to rat pancreatic islet cell apoptosis and insulin secretory defects after prolonged culture in a low non-stimulating glucose concentration. Diabetologia 55:2226–2237
Anello M, Lupi R, Spampinato D et al (2005) Functional and morphological alterations of mitochondria in pancreatic beta cells from type 2 diabetic patients. Diabetologia 48:282–289
Molina AJ, Wikstrom JD, Stiles L et al (2009) Mitochondrial networking protects beta-cells from nutrient-induced apoptosis. Diabetes 58:2303–2315
Dlaskova A, Spacek T, Santorova J et al (2010) 4Pi microscopy reveals an impaired three-dimensional mitochondrial network of pancreatic islet beta-cells, an experimental model of type-2 diabetes. Biochim Biophys Acta 1797:1327–1341
Goulley J, Dahl U, Baeza N, Mishina Y, Edlund H (2007) BMP4-BMPR1A signaling in beta cells is required for and augments glucose-stimulated insulin secretion. Cell Metab 5:207–219
Scott GJ, Ray MK, Ward T et al (2009) Abnormal glucose metabolism in heterozygous mutant mice for a type I receptor required for BMP signaling. Genesis 47:385–391
Bonner C, Farrelly AM, Concannon CG et al (2011) Bone morphogenetic protein 3 controls insulin gene expression and is down-regulated in INS-1 cells inducibly expressing a hepatocyte nuclear factor 1A-maturity-onset diabetes of the young mutation. J Biol Chem 286:25719–25728
Funding
This work was supported by grants from the National Health and Medical Research Council (NHMRC) of Australia. DRL is supported by an Australian Research Council (ARC) Future Fellowship. MB was supported by a Post-doctoral Fellowship from the Société Francophone du Diabète (SFD, Paris, France).
Duality of interest
The authors declare that there is no duality of interest associated with this manuscript.
Contribution statement
MB and DRL conceived and designed experiments, acquired and analysed data and wrote the manuscript. MKM, JL and JYC designed experiments, acquired and analysed data and critically reviewed the manuscript. All authors approved the final version of the manuscript. DRL is responsible for the integrity of the work as a whole.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM Fig. 1
(PDF 260 kb)
ESM Fig. 2
(PDF 224 kb)
ESM Fig. 3
(PDF 181 kb)
ESM Fig. 4
(PDF 267 kb)
ESM Fig. 5
(PDF 270 kb)
ESM Table 1
(PDF 178 kb)
ESM Table 2
(PDF 193 kb)
Rights and permissions
About this article
Cite this article
Bensellam, M., Montgomery, M.K., Luzuriaga, J. et al. Inhibitor of differentiation proteins protect against oxidative stress by regulating the antioxidant–mitochondrial response in mouse beta cells. Diabetologia 58, 758–770 (2015). https://doi.org/10.1007/s00125-015-3503-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00125-015-3503-1