Abstract
Aims/hypothesis
Reduced insulin secretion results in hyperglycaemia and diabetes involving a complex aetiology that is yet to be fully elucidated. Genetic susceptibility is a key factor in beta cell dysfunction and hyperglycaemia but the responsible genes have not been defined. The Collaborative Cross (CC) is a recombinant inbred mouse panel with diverse genetic backgrounds allowing the identification of complex trait genes that are relevant to human diseases. The aim of this study was to identify and characterise genes associated with hyperglycaemia.
Methods
Using an unbiased genome-wide association study, we examined random blood glucose and insulin sensitivity in 53 genetically unique mouse strains from the CC population. The influences of hyperglycaemia susceptibility quantitative trait loci (QTLs) were investigated by examining glucose tolerance, insulin secretion, pancreatic histology and gene expression in the susceptible mice. Expression of candidate genes and their association with insulin secretion were examined in human islets. Mechanisms underlying reduced insulin secretion were studied in MIN6 cells using RNA interference.
Results
Wide variations in blood glucose levels and the related metabolic traits (insulin sensitivity and body weight) were observed in the CC population. We showed that elevated blood glucose in the CC strains was not due to insulin resistance nor obesity but resulted from reduced insulin secretion. This insulin secretory defect was demonstrated to be independent of abnormalities in islet morphology, beta cell mass and pancreatic insulin content. Gene mapping identified the E2f8 (p = 2.19 × 10−15) and Dlg2 loci (p = 3.83 × 10−8) on chromosome 7 to be significantly associated with hyperglycaemia susceptibility. Fine mapping the implicated regions using congenic mice demonstrated that these two loci have independent effects on insulin secretion in vivo. Significantly, our results revealed that increased E2F8 and DLG2 gene expression are correlated with enhanced insulin secretory function in human islets. Furthermore, loss-of-function studies in MIN6 cells demonstrated that E2f8 is involved in insulin secretion through an ATP-sensitive K+ channel-dependent pathway, which leads to a 30% reduction in Abcc8 expression. Similarly, knockdown of Dlg2 gene expression resulted in impaired insulin secretion in response to glucose and non-glucose stimuli.
Conclusions/interpretation
Collectively, these findings suggest that E2F transcription factor 8 (E2F8) and discs large homologue 2 (DLG2) regulate insulin secretion. The CC resource enables the identification of E2f8 and Dlg2 as novel genes associated with hyperglycaemia due to reduced insulin secretion in pancreatic beta cells. Taken together, our results provide better understanding of the molecular control of insulin secretion and further support the use of the CC resource to identify novel genes relevant to human diseases.
Similar content being viewed by others

Introduction
Factors that contribute to beta cell dysfunction in diabetes are heterogeneous and multifactorial, involving a complex interplay between permissive genetic components and detrimental environmental factors [1, 2]. There is ample evidence that diabetes has a strong genetic basis [3, 4], and that islet beta cell dysfunction is inherited [5]. Therefore, understanding the genetic basis of beta cell function will clarify its underlying mechanisms and guide specific therapeutic interventions.
In the past decade, technological advances in genome-wide association studies (GWAS) have led to the discovery of genetic variants marking more than 240 loci implicated in type 2 diabetes risk [6]. Despite the success of human GWAS, current findings account for merely 18% of the overall heritability of type 2 diabetes and most identified variants have no known function and have only a small effect on risk [6, 7]. Moreover, human studies have fundamental limitations that hamper the demonstration of the imputed variants and have confounding environmental factors.
Recombinant inbred mouse populations have been developed as valuable resources with many advantages, including the availability of disease-relevant samples, better control of environmental factors and well-established techniques for genetic modification and functional characterisation of candidate genes [8]. Here, we studied hyperglycaemia in a sophisticated mouse genetic reference population termed the Collaborative Cross (CC). This comprises over 100 genetically diverse inbred strains and was specifically designed for identifying genes mediating complex traits [9, 10]. Each CC strain was derived in a strict breeding programme involving eight parental strains that capture over 90% of the common variation of the mouse genome. The CC resource provides a powerful model system with high genetic diversity, detailed genomic characterisation, balanced allele frequencies and evenly distributed recombination events, which together facilitate high resolution gene mapping and systems genetics studies [11, 12]. The CC mouse resource has been successfully implemented to study complex traits [13,14,15,16,17] and was shown to recapitulate human disease conditions such as diabetic retinopathy [18]. However, few studies have been conducted using the CC resource to study glycaemia-associated traits [19,20,21]. Of these, investigation of gene–environment interactions using CC mice fed a high-fat diet identified a female-specific quantitative trait locus (QTL) (Chr8: 32.02–34.52 Mbp; −log10(p) = 5.9) associated with glucose tolerance [20]. However, the physiological implications of the imputed QTL and putative gene candidates were not explored. The aim of our study was to use the CC resource to identify genes that mediate elevated blood glucose concentrations, and to understand the role of susceptibility genes in islet beta cell function.
Methods
Mice
CC mice were generated and maintained by Geniad (Murdoch, WA, Australia), as detailed by Morahan et al. [10]. All CC mice in this study were at inbreeding generation F15 or beyond with an average of 21 generations. C57BL/6J (B6) mice were purchased from The Walter and Eliza Hall Institute (Kew, VIC, Australia); New Zealand Obese (NZO) mice were bred at the BioResources Facility, Austin Health. All mice were fed ad libitum a standard rodent chow diet (9% fat, 22% protein, and 69% carbohydrate by weight). Congenic mice were generated by mating NZO to B6 mice and backcrossing F1 mice to the B6 parental strain for at least ten generations. Mice from this mating that were heterozygous in the region of interest were selected to commence brother–sister mating to produce B6.NZO-7A and B6.NZO-7C strains. Homozygotes of each line were selected to maintain the subsequent congenic strain. All experimental procedures were performed in accordance with protocols approved by the Austin Health Animal Ethics Committee (AEC No.12/4844 and 14/5196).
Phenotypic screening for metabolic traits
Phenotyping was performed on 1119 mice consisting of 652 males from 53 strains (n = 3–15 mice per strain) and 467 females from 48 strains (n = 3–10 mice per strain) at 8–10 weeks of age. Random blood glucose measurement, ITT and body weight were conducted between 09:00 hours and 11:00 hours. Blood glucose was sampled from tail tipping and measured on an Accu-Chek Performa glucometer (Roche, Switzerland). A non-fasted IPITT was performed as previously described [22].
Genetic analyses
Genome-wide association analysis using the CC resource was performed as previously described [23]. Briefly, mice were genotyped with a MegaMUGA Illumina array (Neogen Genomics, Canada), and HAPPY (http://www.mybiosoftware.com/happy-reconstruct-haplotypes.html) [24] was used to reconstruct founder haplotypes in each strain. Genome-wide association of blood glucose levels was performed using two algorithms: (1) simple GWAS was performed with 77,808 well-annotated SNP markers; (2) linkage analysis identified QTLs and inferred contributing founder haplotypes. A linear mixed model (R/QTL regression) was used to evaluate the maximum-likelihood estimate (derived logarithm of odds [LOD] score) for each genomic position [25]. A significance threshold of 5 × 10−8 was applied. Mapping of QTLs was performed using the GeneMiner software package as described by Ram and Morahan et al. [23, 26]. The confidence interval of each associated region was defined by a 2-LOD drop interval from the peak position. The founder coefficients (LOD scores) for QTLs were presented with the corresponding p value, which was estimated by ANOVA test and reported as −log10(p). All SNPs and genetic loci were positioned with reference to the NCBIm37/mm9 assembly on the UCSC Genome Browser (https://genome.ucsc.edu/).
GTTs, plasma glucose and insulin assays
All GTTs were conducted on male mice at 10–12 weeks of age. OGTTs were performed following a 6 h fast [27]. IVGTTs (1 g/kg) and glucose plus arginine tolerance tests (1 g/kg of each) were performed in overnight fasted mice [28, 29].
Pancreatic histology
Pancreases from B6 and hyperglycaemic strains PIPING and PUB mice were excised and fixed overnight in 4% (wt/vol. in PBS) formaldehyde, dehydrated and embedded in paraffin. Three non-consecutive 5 μm thick sections were sampled from each pancreas (100 μm apart). Sections were stained for insulin using a guinea pig anti-porcine insulin antibody (DAKO, Carpentaria, CA) [30] and counterstained with haematoxylin and eosin. Islet area, number and insulin intensity were analysed with ImageScope software (Aperio Technologies, CA, USA). Beta cell mass was determined as the product of wet pancreas weight and the ratio of insulin positive/total pancreas area.
Pancreatic insulin content
Pancreases from B6, PIPING and PUB mice were examined for insulin content. A portion of mouse pancreas was weighed and homogenised in ice-cold acid–ethanol (0.21 mol/l HCl in 70% ethanol) using an Ultra-Turrax homogeniser (Janke & Kunkel, Staufen, Germany). After overnight incubation at 4°C, the homogenates were centrifuged and supernatants collected. Pellets were resuspended in acid–ethanol for a second extraction. The resulting pancreas extracts were pooled and neutralised prior to assay for insulin by rat Insulin Radioimmunoassay (Millipore, supplied by Abacas, Billerica, MA, USA). Protein concentrations were determined by the Bio-Rad Protein Assay Kit (Bio-Rad, Hercules, CA, USA).
Pancreatic islet isolation and culture
Mouse islets were isolated from B6 and NZO mice as previously described [31, 32]. Mouse islets were cultured in RPMI-1640 medium containing 11.1 mmol/l glucose with 10% FBS, 100 U/ml penicillin and 100 mg/ml streptomycin. For human islet isolation, pancreases were obtained from heart-beating, brain-dead donors (ESM Table 3 and human islet checklist), with consent from next-of-kin and research approval from the St Vincent’s Hospital, Melbourne (HREC-011-04). Human islets were purified using Ficoll density gradients [33] and cultured in Connaught Medical Research Laboratories (CMRL) 1066 medium (Invitrogen, Life Technologies, Carlsbad, CA, USA) supplemented with 10% human serum albumin, 100 U/ml penicillin, 100 mg/ml streptomycin and 2 mmol/l l-glutamine. Stimulation index was measured as how islets respond at 2.8 mmol/l and 28 mmol/l glucose and presented as insulin production at 28 mmol/l divided by insulin production at 2.8 mmol/l in the same islets. All islets were incubated in a 37°C, 5% CO2 humidified incubator.
MIN6 cell culture and gene knockdown
MIN6 cells were kindly provided by Dr. Ross Laybutt, Garvan Institute of Medical Research (Darlinghurst, NSW, Australia) between passage 29 and 36 and cultured in DMEM (Gibco, Life Technologies, Carlsbad, CA, USA) containing 25 mmol/l glucose, 10% (vol./vol.) FBS (Gibco, Life Technologies, Carlsbad, CA, USA), 100 U/ml penicillin and 100 μg/ml streptomycin at 37°C in a humidified incubator. Mycoplasma was tested as negative using LookOut Mycoplasma PCR detection kit (Sigma-Aldrich, St Louis, MO, USA). E2f8 gene knockdown was achieved using Silencer Select small interfering RNA (siRNA; Ambion, Life Technologies, Carlsbad, CA, USA). Scrambled siRNA (Silencer Select Negative control No.1 siRNA) or siRNA duplexes against E2f8 (siRNA ID: s99361) were transfected at 50 nmol/l with 2 μl/ml Lipofectamine RNAiMAX (Invitrogen, Life Technologies, Carlsbad, CA, USA) following the manufacturer’s protocol. Knockdown efficiency was evaluated 72 h post transfection using real-time PCR. Dlg2 knockdown cells were generated via lentiviral-based stable short hairpin RNA (shRNA) transduction according to the manufacturer’s instruction. MISSION lentiviral particles expressing shRNA targeting Dlg2 were manufactured by Sigma-Aldrich (St Louis, MO, USA). Polybrene-treated cells were used as the control for shRNA. All procedures, including lentivirus, were carried out in accordance with protocols approved by the Gene Technology Regulations Committee (NLRD 2016/032).
Insulin secretion assay
Transfected or lentiviral transduced MIN6 cells were cultured in 96-well plates overnight in medium containing 5.5 mmol/l glucose. Secretagogues were prepared in Krebs containing either low (2 mmol/l) or high (20 mmol/l) glucose, 20 mmol/l arginine, 275 μmol/l tolbutamide and 10 nmol/l glucagon-like peptide 1 (GLP-1). KCl-induced insulin secretion was conducted in response to 30 mmol/l KCl with no glucose. Culture medium was collected and assayed using a mouse insulin ELISA (ALPCO Diagnostics, Salem, NH, USA).
RNA extraction, cDNA preparation and real-time quantitative RT-PCR analysis
Total RNA from B6, PIPING and PUB mouse islets was prepared using the RNeasy Mini Kit (Qiagen, Hilden, Germany), MIN6 cells using TRIzol reagent (Ambion, Life Technologies, Carlsbad, CA, USA) and human islets using ISOLATE II RNA Micro Kit (Bioline) according to the manufacturer’s protocol. Purified RNA was quantified by a NanoDrop Lite Spectrophotometer (Thermo Scientific, CA, USA) and treated with DNase using the TURBO DNA-free Kit (Ambion), followed by reverse transcription using SuperScript III First-Strand Synthesis System (Invitrogen). Quantitative real-time PCR was performed with the ViiA 7 real-time PCR thermocycler system using the TaqMan universal PCR master mix reagent kit (Applied Biosystems, Scoresby, Australia) and gene-specific TaqMan Gene Expression Assays (Applied Biosystems) as listed in electronic supplementary material (ESM) Table 1. Gene expression was calculated relative to the housekeeping genes Ins2 for mouse islets, Ppia for MIN6 cells and RPLP0 for human islets. Gene expression was calculated using the ΔΔCt method and reported as fold change of control.
Statistical analysis
All data are presented as mean ± SEM. A Pearson product-moment correlation coefficient was computed to assess the relationship between traits. The AUC was calculated using the trapezoidal rule. Statistical significance between two variables was assessed using unpaired one-tailed Student’s t test. One-way ANOVA with Tukey’s post hoc test was used to determine significant differences between multiple comparisons. For details of statistical significance for genetic analyses, please refer back to the Genetic Analyses section. All analyses were performed using GraphPad Prism 6 (GraphPad Software, La Jolla, CA, USA). Differences between groups were considered statistically significant at the p < 0.05, p < 0.01 and p < 0.001 levels.
Results
Diversity in metabolic traits among the CC mouse population
To characterise the variation in glucose homeostasis in the CC mice, random blood glucose levels were assessed in 1119 mice at 8–10 weeks of age including males from 53 CC strains and females from 48 CC strains (including the founder NZO and B6 strains). A wide range in blood glucose levels was observed: a 2.9-fold (from 7.8 to 22.3 mmol/l) and 2.1-fold (from 6.7 to 14.3 mmol/l) difference among males and females, respectively (Fig. 1a). A higher blood glucose level was noted in males (11.9 ± 0.3 mmol/l vs 9.7 ± 0.2 mmol/l in females; red dashed lines in Fig. 1a). Notably, four strains (PIPING, PUB, DET3 and PEF2) were identified as hyperglycaemic (columns in red), which had blood glucose levels in the upper 2.5% (beyond two standard deviations of the mean) of the tested cohort, regardless of sex.
Phenotypic diversity in metabolic traits among the CC mouse population. (a–c) Random blood glucose level (a), degree of insulin resistance (b; AUC% of basal glucose during ITT) and body weight (c) in males (53 CC strains, n = 3–15 mice per strain) and females (48 CC strains, n = 3–10 mice per strain) at 8–10 weeks of age. Hyperglycaemic strains are highlighted with red columns. The red dashed lines indicate the mean measurements for the traits in males and females, respectively. The reference strain B6 is labelled with grey columns. Data are presented as mean ± SEM. (d) Correlation of blood glucose levels with the degree of insulin resistance among the male mice. Each dot represents a strain. The regression line is shown in red; see also ESM Fig. 1 for further analyses of the CC mouse population
In humans, glucose dysregulation is often driven by insulin resistance and obesity [34]. We therefore determined body weight and insulin sensitivity across 53 strains (Fig. 1b, c). We found that the hyperglycaemic strains had adequate responses to insulin-induced glucose lowering (ESM Fig. 1a, b) and were not overweight (Fig. 1c). In addition, blood glucose levels were negative correlated with the degree of insulin resistance in male mice but not in females (Fig. 1d and Fig. 1c), suggesting hyperglycaemia in male mice was not due to insulin resistance. There was no correlation between blood glucose level and body weights in either sex (ESM Fig. 1d, e). Body weight was positively correlated with insulin resistance in females but not in males (ESM Fig. 1f, g). These results highlight that neither insulin resistance nor obesity was the predominant contributor to hyperglycaemia in this CC population.
Genome-wide association analysis for hyperglycaemia
To identify genetic loci influencing blood glucose concentrations, GWAS and QTL mapping were performed with the random blood glucose data from 53 strains. GWAS revealed four SNPs significantly (p < 5 × 10−8) associated with elevated blood glucose levels in males (Fig. 2a). These SNPs mapped to two unique peaks on chromosome 7 as revealed by QTL analysis (Fig. 2b, Tables 1, 2).
Genome-wide mapping of blood glucose levels identified two loci on chromosome 7. (a) Manhattan plot showing the association of 77,808 SNPs and blood glucose concentration in male mice. The x-axis presents the genomic location and the y-axis shows the significance of each SNP as –log10(p). The thresholds for GWAS significance are indicated as lines drawn at p < 5 × 10−5 (99.9% confidence, blue line) and p < 5 × 10−8 (99.99% confidence, red line). (b) LOD score plot depicting the genomic position of significant peaks for blood glucose level in male mice by linkage analysis. (c) Locus plot (upper) for significant association in male mice at chromosome 7, and founder coefficient plot (lower) illustrating haplotype segregation from eight founder strains on chromosome 7, where high significance is conferred. The red and blue horizontal lines in (b, c) indicate thresholds of 95% and 63% significance, respectively. (d, e) Founder haplotypes in all CC strains at position 52.6–56.7 Mbp (the E2f8 locus) (d) and 98.0–101.5 Mbp (the Dlg2 locus) (e) on chromosome 7. The founder haplotype identity at E2f8 and Dlg2 loci is indicated with colours as shown in the colour scheme. Strains are listed in the order from lower glucose (top) to hyperglycaemic mice (bottom). The strains with NZO haplotypes at each locus had significantly higher blood glucose levels, p = 0.00013 and p = 0.001, respectively (Mann–Whitney U test)
To identify the founder(s) that contributed the causal variants in these hyperglycaemia-susceptible loci, founder haplotype analysis was conducted. There was a strong association of the NZO-derived variants to the hyperglycaemia at both implicated regions on chromosome 7 (Fig. 2c), suggesting that the deleterious effect is attributed to variants derived from the NZO strain. Moreover, the inferred haplotypes for 53 strains showed that the mouse strains that exhibited higher blood glucose levels (PIPING, PUB, DET3 and PEF2) inherited the NZO allele(s) at either one or both implicated loci (Fig. 2d, e).
The most significant locus was located at 52.6–56.7 Mbp on chromosome 7 (LOD = 15.4), a gene-dense region containing more than 65 genes. The peak SNP (rs253243259) showed robust association in independent scans in both male and female mice (p = 2.19 × 10−15 and 9.39 × 10−8, respectively) (Fig. 2a, ESM Fig. 2a). Since the peak SNP (rs253243259) resides in the exon of the gene encoding E2F transcription factor 8 (E2F8; E2f8), this region was designated as the E2f8 locus. A search for candidate genes in this locus based on founder haplotype and sequence analysis identified 37 genes that have variants unique to the NZO mouse genome (ESM Table 2). These variants were predicted to mediate functional alterations in the following genes at the E2f8 locus: Hsd17b14, Snrnp70, Sphk2 and E2f8. Abcc8 and Kcnj11 were also included in the expression analysis due to their known effects on type 2 diabetes.
The other hyperglycaemia susceptibility locus was located approximately 40 Mbp downstream of the E2f8 locus at 98.5–101.5 Mbp (LOD = 8.4). There were seven genes within this region, Dlg2, Ccdc90b, Ankrd42, Pcf11, Ddias, Rab30 and Prcp, although none had protein-changing NZO-unique SNPs. The GWAS identified two significant SNPs at this locus (rs32123098, p = 3.83 × 10−8 and rs243982980, p = 2 × 10−8) both residing in the gene encoding discs large homologue 2 (DLG2; Dlg2), making Dlg2 a high-confidence candidate. This region is hereafter referred to as the Dlg2 locus.
Reduced glucose-stimulated insulin secretion in the hyperglycaemia-susceptible strains
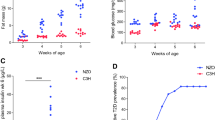
We performed in-depth characterisation in two of the hyperglycaemic strains, PIPING and PUB mice, comparing them with the most commonly used reference B6 strain. The use of PIPING mice allowed us to assess the effect of both NZO-derived E2f8 and Dlg2 alleles, while PUB mice were chosen to determine the individual influence of the detrimental E2f8 allele on glucose homeostasis (Fig. 3a). Both PIPING and PUB mice demonstrated significant hyperglycaemia in fed and fasted states. After an overnight fast, blood glucose levels remained elevated in PUB mice but were normalised in PIPING mice (Fig. 3b). Hyperglycaemia was associated with significantly lower plasma insulin levels in both strains (Fig. 3c), suggesting impaired insulin secretion. The body weights of PIPING and PUB mice were significantly lower compared with B6 mice, regardless of the fasting period (Fig. 3d), although there was no difference in fat pad mass among the three strains (ESM Fig. 3).
Impaired glucose homeostasis and insulin secretion in the hyperglycaemia-susceptible mice. (a) Schematic diagram of haplotype structure at the hyperglycaemia susceptibility loci at Chr7: 51–102 Mbp in PIPING and PUB mice. (b–d) Blood glucose, plasma insulin and body weights in the hyperglycaemic strains, PIPING and PUB, and in B6 mice under non-fasted, 6 h fasted and overnight fasted states (n = 4–17). (e, f) Plasma insulin concentrations and glucose excursions of PIPING (blue squares), PUB (purple triangles) and B6 (black circles) mice following an OGTT (n = 4–14). (g, h) Plasma insulin levels and glucose excursions of PIPING (blue squares), PUB (purple triangles) and B6 (black circles) mice following an IVGTT (n = 6–10). (i) Plasma insulin levels during an intravenous glucose plus arginine tolerance test in PIPING (blue squares), PUB (purple triangles) and B6 (black circles) mice (n = 3–5). Total AUC and incremental AUC (iAUC) of glucose and insulin during GTTs are presented as corresponding panel inserts in (e, f, g, i). Data are presented as mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001 for PIPING vs B6; †p < 0.05, ††p < 0.01, †††p < 0.001 for PUB vs B6. O/N, overnight
OGTTs revealed significant reduction in insulin secretion in the hyperglycaemia-susceptible strains when compared with B6 mice (Fig. 3e), which resulted in glucose intolerance (Fig. 3f). Peak insulin secretion was decreased in PIPING and PUB mice compared with B6 mice (Fig. 3g), despite a greater level of glucose stimulus (Fig. 3h). Plasma insulin excursions following a bolus of arginine plus glucose demonstrated a substantial decline in insulin secretion in both hyperglycaemic strains, particularly in PUB mice (Fig. 3i).
To further determine whether these two loci have independent effects on insulin secretion, the NZO-derived E2f8 and Dlg2 alleles were bred onto the B6 genetic background, creating two congenic mouse strains. As shown in ESM Fig. 4a–c, the NZO-derived E2f8 and Dlg2 alleles account for a substantial decline in insulin secretion as revealed in B6.NZO-7A mice, while a significant improvement only in first-phase insulin secretion was found in B6.NZO-7C congenic mice bearing only the NZO allele at Dlg2 locus (ESM Fig. 4b–c) [35]. Importantly, the insulin secretory defects persisted in the B6.NZO-7C mice with the NZO Dlg2 allele, demonstrated by a greater than fivefold reduction in tolbutamide-induced early-phase insulin secretion when compared with B6, suggesting an appreciable effect of the NZO-Dlg2 allele alone on insulin secretion. These results demonstrate that E2f8 and Dlg2 loci have substantial and independent effects on insulin secretion in vivo, with potent genetic determinants predisposing to beta cell dysfunction and subsequent hyperglycaemia.
Morphological and histological examination of pancreatic beta cells
Pancreatic histological analysis revealed that both PIPING and PUB mice had comparable numbers of islets to B6 mice (Fig. 4a). Bigger islets (Fig. 4b) and increased islet proportion (Fig. 4c) were identified in PIPING and PUB mice, which was associated with a significant increase in the proportion of large islets (Fig. 4d). Beta cell mass (Fig. 4e) and pancreatic insulin content (Fig. 4f) were not reduced in the hyperglycaemic strains. These data suggest the hyperglycaemia in PUB and PIPING strains was due primarily to functional defects in insulin secretion rather than beta cell loss.
Normal pancreatic histology and insulin content in the hyperglycaemic strains. (a–c) Number, size and proportion of islet in 24-week-old PIPING, PUB and B6 mice as examined by immunohistochemistry (IHC). Images shown are representative of IHC for B6, PIPING and PUB mice. (d) Proportion of islet size (small, medium and large) is presented as percentage of total islet number. (e, f) Beta cell mass and pancreatic insulin content (proportion to total protein content) in PIPING, PUB and B6 mice at 24 weeks of age. Data are presented as mean ± SEM (n = 4–7). *p < 0.05, **p < 0.01 for PIPING vs B6; †p < 0.05, ††p < 0.01 for PUB vs B6
Identification and validation of E2f8 and Dlg2 as islet genes that regulate pancreatic insulin secretion
To determine whether these hyperglycaemia-susceptible NZO alleles contribute to beta cell dysfunction, expression of the candidate genes was determined in primary islets from diabetes-susceptible NZO mice compared with B6 mice. At the E2f8 locus, Abcc8, Kcnj11 and E2f8 expression were significantly reduced in NZO islets, while there was no difference in Snrnp70 and Sphk2 (Fig. 5a). No Hsd17b14 transcripts were detected. Although the roles of Abcc8 and Kcnj11 in insulin secretion are well established, the reduced E2f8 expression in diabetes-susceptible islets is of interest and novel, so was further characterised.
E2f8 and Dlg2 knockdowns resulted in impaired insulin secretion in MIN6 cells. (a) Real-time PCR analysis of islet mRNA expression of candidate genes at Chr7: 52.6–56.7 Mbp in B6 and NZO male mice. (b) Islet mRNA expression of all genes at Chr7: 98.0–101.5 Mbp in isolated islets from B6 and NZO male mice. Relative mRNA expression was calculated relative to an islet housekeeping gene, Ins2. Data are shown as fold change relative to B6 levels for each gene (\( {2}^{\Delta \Delta {\mathrm{C}}_{\mathrm{t}}} \) ± \( {2}^{\Delta \Delta {\mathrm{C}}_{\mathrm{t}}\pm \mathrm{SEM}} \)) and presented as mean ± SEM (n = 4–6). In (a, b) *p < 0.05 vs B6. (c, d) Correlation of human E2F8 and DLG2 gene expression with insulin stimulation index in human pancreatic islets from healthy donors. Each dot presents the data from one donor. The regression line is shown in red. (e) E2f8 mRNA expression by real-time PCR in MIN6 cells at 72 h post transfection of scramble or E2f8 siRNA. (f) Insulin secretion from scramble or E2f8 siRNA transfected MIN6 cells at basal (2 mmol/l glucose) and high glucose (20 mmol/l). (g–i) Insulin secretion in response to secretagogues (275 μmol/l tolbutamide, 20 mmol/l arginine and 10 nmol/l GLP-1) at basal (2 mmol/l glucose) or high glucose (20 mmol/l), and (j) in response to KCl (30 mmol/l) with no glucose, in MIN6 cells transfected with scramble or E2f8 siRNA. Control (Ctrl) in (j) is basal glucose alone. (k, l) Abcc8 and Kcnj11 mRNA expression in scramble or E2f8 siRNA transfected MIN6 cells. (m) Dlg2 mRNA expression by real-time PCR in MIN6 cells with lenti-Dlg2 shRNA stable transduction (Dlg2-shRNA) or polybrene-treated control. (n) Insulin secretion from Dlg2-shRNA and polybrene-treated MIN6 cells at basal (2 mmol/l glucose) and high glucose (20 mmol/l). (o–q) Insulin secretion in response to secretagogues (275 μmol/l tolbutamide, 20 mmol/l arginine and 10 nmol/l GLP-1) at basal (2 mmol/l glucose) or high glucose (20 mmol/l), and (r) in response to KCl (30 mmol/l) with no glucose, in Dlg2-shRNA and polybrene-treated MIN6 cells. Control in (r) is basal glucose alone. Relative mRNA expression in (e, k, l, m) was determined relative to the housekeeping gene, Ppia. Data are presented as mean ± SEM, n = 3–11 per group, three replicates in each group. *p < 0.05, **p < 0.01, ***p < 0.001 as shown
Interestingly, at the Dlg2 locus, significant reductions in Ddias and Ccdc90b were observed in NZO islets, whereas Dlg2 (p = 0.065), Ankrd42, Pcf11, Rab30 and Prcp expression were comparable between groups (Fig. 5b).
Expression of candidate genes in human islets
Human pancreatic islets from seven diabetic and eight non-diabetic donors were examined for candidate gene expression (ESM Table 3). There was no significant difference in any of the candidate genes (ESM Fig. 5). Despite the low E2F8 expression (mean Ct of 36.4 ± 0.5 vs 23.5 ± 0.5 in RPLP0) in human islets, increased E2F8 and DLG2 gene expression was noted to be significantly correlated with enhanced beta cell function in non-diabetic donors (Fig. 5c, d), suggesting these genes have important roles in insulin secretion.
Effect of downregulation of E2f8 and Dlg2 on beta cell insulin secretion
To validate whether E2f8 and Dlg2 are involved in insulin secretory function, RNA interference (RNAi) was utilised to reduce their expression in MIN6 cells. A greater than 50% knockdown of E2f8 expression led to impaired glucose-stimulated insulin secretion (1116.6 ± 167.5 at 2 mmol/l vs 1526.4 ± 384.4 pmol/l at 20 mmol/l in E2f8 siRNA group, p = 0.178; 1.3 ± 0.3-fold insulin secretion from 2 mmol/l to 20 mmol/l in E2f8 knockdown cells compared with 2.2 ± 0.2 fold in the scramble control, p = 0.044). (Fig. 5e, f).
We next investigated insulin secretion in response to a series of non-glucose secretagogues, such as tolbutamide, an ATP-sensitive K+ (KATP) channel antagonist, and arginine, which causes membrane depolarisation. Results showed that tolbutamide- and arginine-induced insulin secretion were attenuated in E2f8 siRNA-treated cells compared with scramble siRNA control (Fig. 5g, h). Interestingly, incretin (GLP-1)-stimulated insulin secretion at 20 mmol/l glucose in the E2f8 knockdown cells was comparable to the scramble control (Fig. 5i). KCl-induced insulin secretion from E2f8 knockdown cells was no different from control (Fig. 5j). Abcc8 was significantly decreased by 30% in E2f8 knockdown cells, while Kcnj11 remained unaltered (Fig. 5k, l). E2F8 is a known cell cycle regulator, therefore apoptotic markers Bcl2, Ddit3 and Bak1 were assessed and revealed no effects in the E2f8 knockdown cells (ESM Fig. 6a-c). These results indicate a previously unexplored role of E2F8 in insulin secretory function via regulating the KATP channel in pancreatic beta cells.
Dlg2 expression was reduced by 60% using shRNA in MIN6 cells (Fig. 5m). Insulin secretion in response to 20 mmol/l glucose as well as tolbutamide, arginine, GLP-1 and KCl was markedly reduced in Dlg2 knockdown cells (Fig. 5n–r), indicating Dlg2 is involved in insulin secretory function. Dlg2 has been implicated in cell death pathways through glutamate receptors [36]; however, our results demonstrated the reduced insulin secretion in Dlg2 knockdown cells had no effect on expression of cell death genes Bcl2 and Ddit3 (ESM Fig. 6d, e).
Discussion
The aims of this study were to identify genes influencing blood glucose levels using a genetically diverse mouse population and to demonstrate the biological significance of these regions in the pathogenesis of hyperglycaemia. Using the CC resource, we identified two loci on chromosome 7 significantly associated with hyperglycaemia. The genetic lesions led to substantial defects in insulin secretion and, in turn, to overt glucose intolerance. Furthermore, we identified and characterised E2f8 and Dlg2 as genes contributing to hyperglycaemia and beta cell dysfunction. The power of the CC resource is shown by its ability to rapidly identify candidate genes, and even candidate nucleotide changes, that impact on complex traits such as hyperglycaemia.
The E2f8 locus was identified proximal to a previously reported region (Chr7: 56.2–62.5 Mbp) for plasma glucose in an independent mouse population [37]. In addition, human GWAS have reported associations with type 2 diabetes [7, 38,39,40,41], cholesterol [42, 43] and obesity-related traits [44] in the human orthologue region. At the E2f8 locus, previous studies were primarily focused on Abcc8 (encoding sulfonylurea receptor 1 [SUR1]) and Kcnj11 (encoding inwardly-rectifying K+ channel 6.2 [Kir6.2]), both components of the KATP channel [39, 40]. In line with our previous study, we identified decreased Abcc8 and Kcnj11 expression in NZO islets, which was attributable to the insertion/deletion events in the NZO Abcc8 allele (within the E2f8 locus) [35]. Interestingly, replacement with a functional Abcc8 gene restored only the early-phase insulin secretion in transgenic NZO mice, suggesting that other genes within the candidate region may also be involved (ESM Fig. 4). The present study revealed a reduction in a novel gene, E2f8, in diabetes-susceptible NZO islets, which contributed to impaired insulin secretion and reduced Abcc8 expression in MIN6 cells. Our results indicate that multiple genetic variants in NZO alleles in this region contribute to insulin secretion by altering the expression or function of several effector transcripts. Moreover, the present study showed that PIPING and PUB mice have defects in both phases of insulin secretion in response to glucose and arginine, whereas we previously showed that NZO mice exhibited reduced insulin secretion in response to glucose and tolbutamide but not to arginine [35]. Together, these observations led us to postulate that E2f8 and Dlg2 genes take part in the late phase of insulin secretion and suggest substantial and diverse effects of these deleterious NZO alleles in mice with divergent genetic backgrounds.
E2F8, a member of E2F transcription factor family, has a well-established role in cell cycle regulation and tumourigenesis [45]. An increase in E2f8 mRNA expression in subcutaneous adipose tissue was associated with impaired glucose tolerance in obese individuals [46]. In addition, E2f8 expression was markedly increased in response to high-fat feeding and glucose treatment in mice and in an adipocyte-derived cell line [47, 48]. These studies suggested E2F8 was associated with impaired glucose tolerance and obesity, states known to cause insulin hypersecretion.
Our study identified a nonsynonymous polymorphism (rs253243259) that was strongly associated with hyperglycaemia in GWAS. This SNP in exon 8 of E2f8 results in an amino acid substitution from positively charged lysine to a polar neutral asparagine (Lys401Asn) in the GAP-43-like domain; however, the biological function of this variant and E2F8 in glucose homeostasis has not been reported previously. Furthermore, we identified that increased E2F8 expression is positively correlated with enhanced stimulation index in human islets. Our study provides compelling evidence for E2F8 as a novel effector in beta cell function and ABCC8 as a potential regulatory target of E2F8.
A previous study of islet gene profiling [49] showed that changes in cell cycle-related genes including E2f8 predicted diabetes susceptibility through modulating beta cell proliferation. Although previous studies have suggested a number of cell cycle-related genes may modulate beta cell fate, we showed a lower E2f8 mRNA expression in diabetes-susceptible NZO islets, and this reduction leads to reduced insulin secretion without affecting cell survival, as evidenced by no change in expression of genes involved in apoptosis. Our results suggest a novel role of E2F8 in insulin secretion independent of cell cycle regulation. In support of this finding, increasing evidence has suggested non-canonical functions of cell cycle genes in metabolic processes, such as insulin secretion [50], hepatic glucose production [51] and adipogenesis [52,53,54]. For example, E2F1, a pivotal activator and regulatory target of E2F8 [55], has been shown to control insulin secretion through transcriptional regulation of Kcnj11 (Kir6.2) gene expression [50] (Fig. 6). It is worth noting that E2f8 levels were downregulated (28.6-fold) in E2f1−/− mouse islets [50], implying a potential role of E2F8 in an insulin deficient mouse model. More importantly, our results demonstrate that reduced insulin secretion in E2f8 knockdown cells is specific to glucose, tolbutamide and arginine but not GLP-1 or KCl, indicating that E2F8 regulates insulin secretion through a KATP-dependent signalling pathway. In addition, Abcc8 was significantly decreased by 30% in E2f8 knockdown cells. Our results present the first direct evidence for E2F8 as a novel effector of insulin secretion, potentially through the regulation of Abcc8 gene expression.
Putative roles of E2F8 and DLG2 in glucose-stimulated insulin secretion in the pancreatic beta cells. Glucose-stimulated insulin secretion initiates from glucose uptake by pancreatic beta cells through the GLUT2 glucose transporter; subsequently the intracellular glucose is phosphorylated by glucokinase (GK) then oxidised to generate ATP. The increased ATP/ADP ratio results in the closure of the KATP channel, which triggers membrane depolarisation, and in turn activates the voltage-dependent Ca2+ channel (VDCC), which elicits the exocytosis of insulin granules. E2F8 may be involved in this machinery through alteration of the KATP channel by directly changing Abcc8 (SUR1) gene expression, and/or regulating E2F1 at the transcriptional level, the reduced E2F1 leading to decreased expression of Kcnj11 (Kir6.2). On the other hand, DLG2 acting on islet NMDAR ensures proper function leading to beta cell membrane repolarisation, thus opening the KATP channel. This figure was created in BioRender.com
Our GWAS also identified two SNPs (rs32123098 and rs243982980) in the Dlg2 gene as being associated with hyperglycaemia. Dlg2 encodes postsynaptic density protein (PSD)-93, which forms scaffold complexes for clustering and anchoring membrane proteins, such as ionotropic N-methyl-d-aspartate receptors (NMDARs) [56] and K+ channel clusters [57]. A recent study reported that NMDAR in islets was involved in the regulation of insulin release and blood glucose control [58]. In addition, NMDARs and K+ channels have been suggested to be drug targets to regulate pancreatic beta cell function for treating insulin-dependent diabetes [58, 59]. The human DLG2 locus was associated with the disposition index, an indicator of beta cell function, in a small African-American cohort [60]. Furthermore, the Dlg2 gene has been associated with height and BMI as well as type 2 diabetes and glycaemic traits [61]. In the present study, our results indicated genetic ablation of Dlg2 in MIN6 cells resulted in a generalised defect in insulin secretion. Of interest, our previous study showed the NZO-derived Dlg2 allele has an independent effect on insulin secretion as demonstrated by a greater than fivefold reduction in tolbutamide-induced early-phase insulin secretion in a congenic strain carrying the NZO-derived Dlg2 allele on a B6 background (ESM Fig. 4) [35]. These results support our hypothesis that Dlg2 participates in pancreatic insulin secretory function.
The concept of precision medicine in diabetes has been considered as a more effective strategy than conventional treatments [62]. However, the application to type 2 diabetes has been challenging owing to its complex polygenic pathogenesis, and hence new approaches to improve our understanding of the genetic basis of glucose regulation are critical. Our study provides proof-of-principle work of rapid gene identification for the complex traits of insulin secretion and blood glucose concentrations, and highlights roles for E2f8 and Dlg2 in islet function and glucose homeostasis. Future studies on profiling the differential response to a series of glucose-lowering drugs in the CC mouse population will perhaps facilitate the understanding of the biological role of E2F8 and DLG2 in clinical applications in humans.
Data availability statement
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- B6:
-
C57BL/6J
- CC:
-
Collaborative Cross
- DLG2:
-
Discs large homologue 2
- E2F8:
-
E2F transcription factor 8
- GLP-1:
-
Glucagon-like peptide 1
- GWAS:
-
Genome-wide association study
- KATP channel:
-
ATP-sensitive K+ channel
- LOD:
-
Logarithm of odds
- NMDAR:
-
Ionotropic N-methyl-d-aspartate receptor
- NZO:
-
New Zealand Obese
- QTL:
-
Quantitative trait locus
- shRNA:
-
Short hairpin RNA
- siRNA:
-
Small interfering RNA
References
Andrikopoulos S (2010) Obesity and type 2 diabetes: slow down!—can metabolic deceleration protect the islet beta cell from excess nutrient-induced damage? Mol Cell Endocrinol 316(2):140–146. https://doi.org/10.1016/j.mce.2009.09.031
DeFronzo RA (2004) Pathogenesis of type 2 diabetes mellitus. Med Clin North Am 88(4):787–835. https://doi.org/10.1016/j.mcna.2004.04.013
Hemminki K, Li X, Sundquist K, Sundquist J (2010) Familial risks for type 2 diabetes in Sweden. Diabetes Care 33(2):293–297. https://doi.org/10.2337/dc09-0947
Meigs JB, Cupples LA, Wilson PW (2000) Parental transmission of type 2 diabetes: the Framingham Offspring Study. Diabetes 49(12):2201–2207. https://doi.org/10.2337/diabetes.49.12.2201
Elbein SC, Hasstedt SJ, Wegner K, Kahn SE (1999) Heritability of pancreatic β-cell function among nondiabetic members of Caucasian familial type 2 diabetic kindreds. J Clin Endocrinol Metab 84(4):1398–1403. https://doi.org/10.1210/jcem.84.4.5604
Mahajan A, Taliun D, Thurner M et al (2018) Fine-mapping type 2 diabetes loci to single-variant resolution using high-density imputation and islet-specific epigenome maps. Nat Genet 50(11):1505–1513. https://doi.org/10.1038/s41588-018-0241-6
DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium, Asian Genetic Epidemiology Network Type 2 Diabetes (AGEN-T2D) Consortium, South Asian Type 2 Diabetes (SAT2D) Consortium et al (2014) Genome-wide trans-ancestry meta-analysis provides insight into the genetic architecture of type 2 diabetes susceptibility. Nat Genet 46(3):234–244
Skarnes WC, Rosen B, West AP et al (2011) A conditional knockout resource for the genome-wide study of mouse gene function. Nature 474(7351):337–342. https://doi.org/10.1038/nature10163
Churchill GA, Airey DC, Allayee H et al (2004) The Collaborative Cross, a community resource for the genetic analysis of complex traits. Nat Genet 36(11):1133–1137. https://doi.org/10.1038/ng1104-1133
Morahan G, Balmer L, Monley D (2008) Establishment of “The Gene Mine”: a resource for rapid identification of complex trait genes. Mamm Genome 19(6):390–393. https://doi.org/10.1007/s00335-008-9134-9
Aylor DL, Valdar W, Foulds-Mathes W et al (2011) Genetic analysis of complex traits in the emerging Collaborative Cross. Genome Res 21(8):1213–1222. https://doi.org/10.1101/gr.111310.110
Collaborative Cross Consortium (2012) The genome architecture of the Collaborative Cross mouse genetic reference population. Genetics 190(2):389–401. https://doi.org/10.1534/genetics.111.132639
Ferguson B, Ram R, Handoko HY et al (2015) Melanoma susceptibility as a complex trait: genetic variation controls all stages of tumor progression. Oncogene 34(22):2879–2886. https://doi.org/10.1038/onc.2014.227
Nachshon A, Abu-Toamih Atamni HJ, Steuerman Y et al (2016) Dissecting the effect of genetic variation on the hepatic expression of drug disposition genes across the Collaborative Cross mouse strains. Front Genet 7:172
Boutilier JK, Taylor RL, Ram R et al (2017) Variable cardiac α-actin (Actc1) expression in early adult skeletal muscle correlates with promoter methylation. Biochim Biophys Acta 1860(10):1025–1036. https://doi.org/10.1016/j.bbagrm.2017.08.004
Chitsazan A, Ferguson B, Ram R et al (2016) A mutation in the Cdon gene potentiates congenital nevus development mediated by NRAS(Q61K). Pigment Cell Melanoma Res 29(4):459–464. https://doi.org/10.1111/pcmr.12487
Chick JM, Munger SC, Simecek P et al (2016) Defining the consequences of genetic variation on a proteome-wide scale. Nature 534(7608):500–505. https://doi.org/10.1038/nature18270
Weerasekera LY, Balmer LA, Ram R, Morahan G (2015) Characterization of retinal vascular and neural damage in a novel model of diabetic retinopathy. Invest Ophthalmol Vis Sci 56(6):3721–3730. https://doi.org/10.1167/iovs.14-16289
Nashef A, Abu-Toamih Atamni HJ, Buchnik Y et al (2017) Collaborative cross mouse population for studying alveolar bone changes and impaired glucose tolerance comorbidity after high-fat diet consumption. J Periodontol 88(9):e150–e158. https://doi.org/10.1902/jop.2017.170075
Abu-Toamih Atamni HJ, Ziner Y, Mott R, Wolf L, Iraqi FA (2017) Glucose tolerance female-specific QTL mapped in collaborative cross mice. Mamm Genome 28(1–2):20–30. https://doi.org/10.1007/s00335-016-9667-2
Atamni HJ, Mott R, Soller M, Iraqi FA (2016) High-fat-diet induced development of increased fasting glucose levels and impaired response to intraperitoneal glucose challenge in the collaborative cross mouse genetic reference population. BMC Genet 17(1):10. https://doi.org/10.1186/s12863-015-0321-x
Lamont BJ, Visinoni S, Fam BC et al (2006) Expression of human fructose-1,6-bisphosphatase in the liver of transgenic mice results in increased glycerol gluconeogenesis. Endocrinology 147(6):2764–2772. https://doi.org/10.1210/en.2005-1498
Ram R, Mehta M, Balmer L, Gatti DM, Morahan G (2014) Rapid identification of major-effect genes using the collaborative cross. Genetics 198(1):75–86. https://doi.org/10.1534/genetics.114.163014
Mott R, Talbot CJ, Turri MG, Collins AC, Flint J (2000) A method for fine mapping quantitative trait loci in outbred animal stocks. Proc Natl Acad Sci U S A 97(23):12649–12654. https://doi.org/10.1073/pnas.230304397
Broman KW, Sen S (2009) A guide to QTL mapping with R/qtl. Springer, New York; London
Ram R, Mehta M, Nguyen QT et al (2016) Systematic evaluation of genes and genetic variants associated with type 1 diabetes susceptibility. J Immunol 196(7):3043–3053. https://doi.org/10.4049/jimmunol.1502056
Andrikopoulos S, Blair AR, Deluca N, Fam BC, Proietto J (2008) Evaluating the glucose tolerance test in mice. Am J Physiol Endocrinol Metab 295(6):e1323–e1332. https://doi.org/10.1152/ajpendo.90617.2008
Kebede M, Favaloro J, Gunton JE et al (2008) Fructose-1,6-bisphosphatase overexpression in pancreatic β-cells results in reduced insulin secretion: a new mechanism for fat-induced impairment of β-cell function. Diabetes 57(7):1887–1895. https://doi.org/10.2337/db07-1326
Andrikopoulos S, Massa CM, Aston-Mourney K et al (2005) Differential effect of inbred mouse strain (C57BL/6, DBA/2, 129T2) on insulin secretory function in response to a high fat diet. J Endocrinol 187(1):45–53. https://doi.org/10.1677/joe.1.06333
Lamont BJ, Drucker DJ (2008) Differential antidiabetic efficacy of incretin agonists versus DPP-4 inhibition in high fat fed mice. Diabetes 57(1):190–198. https://doi.org/10.2337/db07-1202
Zraika S, Dunlop M, Proietto J, Andrikopoulos S (2002) The hexosamine biosynthesis pathway regulates insulin secretion via protein glycosylation in mouse islets. Arch Biochem Biophys 405(2):275–279. https://doi.org/10.1016/s0003-9861(02)00397-1
Aston-Mourney K, Wong N, Kebede M et al (2007) Increased nicotinamide nucleotide transhydrogenase levels predispose to insulin hypersecretion in a mouse strain susceptible to diabetes. Diabetologia 50(12):2476–2485. https://doi.org/10.1007/s00125-007-0814-x
Ricordi C, Lacy PE, Finke EH, Olack BJ, Scharp DW (1988) Automated method for isolation of human pancreatic islets. Diabetes 37(4):413–420. https://doi.org/10.2337/diab.37.4.413
Kahn BB, Flier JS (2000) Obesity and insulin resistance. J Clin Invest 106(4):473–481. https://doi.org/10.1172/JCI10842
Andrikopoulos S, Fam BC, Holdsworth A et al (2016) Identification of ABCC8 as a contributory gene to impaired early-phase insulin secretion in NZO mice. J Endocrinol 228(1):61–73. https://doi.org/10.1530/JOE-15-0290
Wu HC, Chen CM, Chen YC, Fung HC, Chang KH, Wu YR (2018) DLG2, but not TMEM229B, GPNMB, and ITGA8 polymorphism, is associated with Parkinson's disease in a Taiwanese population. Neurobiol Aging 64:158 e1-158.e6
Parks BW, Sallam T, Mehrabian M et al (2015) Genetic architecture of insulin resistance in the mouse. Cell Metab 21(2):334–346. https://doi.org/10.1016/j.cmet.2015.01.002
Scott LJ, Mohlke KL, Bonnycastle LL et al (2007) A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science 316(5829):1341–1345. https://doi.org/10.1126/science.1142382
Zeggini E, Scott LJ, Saxena R et al (2008) Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat Genet 40(5):638–645. https://doi.org/10.1038/ng.120
Zeggini E, Weedon MN, Lindgren CM et al (2007) Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science 316(5829):1336–1341. https://doi.org/10.1126/science.1142364
Diabetes Genetics Initiative of Broad Institute of Harvard and MIT, Lund University, and Novartis Institutes of BioMedical Research, Saxena R, Voight BF et al (2007) Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science 316(5829):1331–1336. https://doi.org/10.1126/science.1142358
Teslovich TM, Musunuru K, Smith AV et al (2010) Biological, clinical and population relevance of 95 loci for blood lipids. Nature 466(7307):707–713. https://doi.org/10.1038/nature09270
Global Lipids Genetics Consortium, Willer CJ, Schmidt EM et al (2013) Discovery and refinement of loci associated with lipid levels. Nat Genet 45(11):1274–1283. https://doi.org/10.1038/ng.2797
Locke AE, Kahali B, Berndt SI et al (2015) Genetic studies of body mass index yield new insights for obesity biology. Nature 518(7538):197–206. https://doi.org/10.1038/nature14177
Maiti B, Li J, de Bruin A et al (2005) Cloning and characterization of mouse E2F8, a novel mammalian E2F family member capable of blocking cellular proliferation. J Biol Chem 280(18):18211–18220. https://doi.org/10.1074/jbc.M501410200
Minchenko OH, Bashta YM, Minchenko DO, Ratushna OO (2016) Glucose tolerance in obese men is associated with dysregulation of some angiogenesis-related gene expressions in subcutaneous adipose tissue. Fiziol Zh 62(2):12–23. https://doi.org/10.15407/fz62.02.012
Suwa A, Yoshino M, Yamazaki C et al (2010) RMI1 deficiency in mice protects from diet and genetic-induced obesity. FEBS J 277(3):677–686. https://doi.org/10.1111/j.1742-4658.2009.07513.x
Suwa A, Kurama T, Shimokawa T (2011) Adipocyte hyperplasia and RMI1 in the treatment of obesity. FEBS J 278(4):565–569. https://doi.org/10.1111/j.1742-4658.2010.07980.x
Keller MP, Choi Y, Wang P et al (2008) A gene expression network model of type 2 diabetes links cell cycle regulation in islets with diabetes susceptibility. Genome Res 18(5):706–716. https://doi.org/10.1101/gr.074914.107
Annicotte JS, Blanchet E, Chavey C et al (2009) The CDK4-pRB-E2F1 pathway controls insulin secretion. Nat Cell Biol 11(8):1017–1023. https://doi.org/10.1038/ncb1915
Lee Y, Dominy JE, Choi YJ et al (2014) Cyclin D1-Cdk4 controls glucose metabolism independently of cell cycle progression. Nature 510(7506):547–551. https://doi.org/10.1038/nature13267
Hanse EA, Mashek DG, Becker JR et al (2012) Cyclin D1 inhibits hepatic lipogenesis via repression of carbohydrate response element binding protein and hepatocyte nuclear factor 4α. Cell Cycle 11(14):2681–2690. https://doi.org/10.4161/cc.21019
Abella A, Dubus P, Malumbres M et al (2005) Cdk4 promotes adipogenesis through PPARγ activation. Cell Metab 2(4):239–249. https://doi.org/10.1016/j.cmet.2005.09.003
Hydbring P, Malumbres M, Sicinski P (2016) Non-canonical functions of cell cycle cyclins and cyclin-dependent kinases. Nat Rev Mol Cell Biol 17(5):280–292. https://doi.org/10.1038/nrm.2016.27
Li J, Ran C, Li E et al (2008) Synergistic function of E2F7 and E2F8 is essential for cell survival and embryonic development. Dev Cell 14(1):62–75. https://doi.org/10.1016/j.devcel.2007.10.017
Kim E, Cho KO, Rothschild A, Sheng M (1996) Heteromultimerization and NMDA receptor-clustering activity of Chapsyn-110, a member of the PSD-95 family of proteins. Neuron 17(1):103–113. https://doi.org/10.1016/s0896-6273(00)80284-6
Leyland ML, Dart C (2004) An alternatively spliced isoform of PSD-93/chapsyn 110 binds to the inwardly rectifying potassium channel, Kir2.1. J Biol Chem 279(42):43427–43436. https://doi.org/10.1074/jbc.M407575200
Marquard J, Otter S, Welters A et al (2015) Characterization of pancreatic NMDA receptors as possible drug targets for diabetes treatment. Nat Med 21(4):363–372. https://doi.org/10.1038/nm.3822
Choi BH, Hahn SJ (2010) Kv1.3: a potential pharmacological target for diabetes. Acta Pharmacol Sin 31(9):1031–1035. https://doi.org/10.1038/aps.2010.133
Palmer ND, Mychaleckyj JC, Langefeld CD et al (2010) Evaluation of DLG2 as a positional candidate for disposition index in African-Americans from the IRAS Family Study. Diabetes Res Clin Pract 87(1):69–76. https://doi.org/10.1016/j.diabres.2009.10.015
Type 2 Diabetes Knowledge Portal: DLG2. 2015; Available from http://www.type2diabetesgenetics.org/gene/geneInfo/DLG2. Accessed 1 Dec 2019
McCarthy MI (2017) Painting a new picture of personalised medicine for diabetes. Diabetologia 60(5):793–799. https://doi.org/10.1007/s00125-017-4210-x
Acknowledgements
Thanks to R. Sgambellone and F. Manippa for technical assistance and contribution to the phenotypic screening of the CC mice. We thank Geniad Pty Ltd. for providing the CC mice used in this study. We are grateful to R. Ram for supporting the genetic analysis. We thank all organ donors and their families, Donatelife and the staff of St Vincent’s Institute for the human islets.
Funding
This work was supported by Diabetes Australia Research Program, the Operational Infrastructure Support Scheme of the Government of Victoria, and the Diabetes Research Foundation of Western Australia.
Author information
Authors and Affiliations
Contributions
CY, SPM, HET, GM and SA conceived the presented idea and designed the experiments. CY, GM and SA verified the analytical methods. GM designed the model and the computational framework. CY, SPM and MW carried out the experiment and analysed the data. CY, SPM, MW, TL, KL, HET, GM and SA contributed to sample acquisition and preparation. CY, SPM, TL, KL, HET, GM and SA contributed to the analysis and interpretation of the results. CY wrote the manuscript with support from GM and SA. CY, SPM, MW, TL, KL, HET, GM and SA provided critical feedback, helped shape the research, and reviewed and revised the manuscript. CY, SPM, MW, TL, KL, HET, GM and SA contributed to the final approval of the version to be published. CY is responsible for the integrity of the work as a whole.
Corresponding authors
Ethics declarations
The authors declare that there are no relationships or activities that may bias, or be perceived to bias, this work.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Grant Morahan and Sofianos Andrikopoulos jointly led this work.
Rights and permissions
About this article
Cite this article
Yang, CH., Mangiafico, S.P., Waibel, M. et al. E2f8 and Dlg2 genes have independent effects on impaired insulin secretion associated with hyperglycaemia. Diabetologia 63, 1333–1348 (2020). https://doi.org/10.1007/s00125-020-05137-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00125-020-05137-0