Abstract
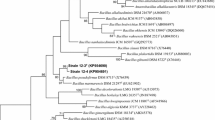
The taxonomic position of a Gram-positive, endospore-forming bacterium isolated from soil sample collected from an industrial site was analyzed by a polyphasic approach. The strain designated as IITR-54T matched most of the phenotypic and chemical characteristics of the genus Bacillus and represents a novel species. It was found to biodegrade 4-chlorobiphenyl through dechlorination and was isolated through enrichment procedure from an aged polychlorinated biphenyl-contaminated soil. Both resting cell assay and growth under aerobic liquid conditions using 4-chlorobiphenyl as sole source of carbon along with 0.01 % yeast extract, formation of chloride ions was measured. 16S rRNA (1,489 bases) nucleotide sequence of isolated strain was compared with those of closely related Bacillus type strains and confirmed that the strain belongs to the genus Bacillus. Strain IITR-54T differs from all other species of Bacillus by at least 2.1 % at the 16S rRNA level, and the moderately related species are Bacillus oceanisediminis (97.9 %) followed by Bacillus infantis (97.7 %), Bacillus firmus (97.4 %), Bacillus drentensis (97.3 %), Bacillus circulans (97.2 %), Bacillus soli (97.1 %), Bacillus horneckiae (97.1 %), Bacillus pocheonensis (97.1 %) and Bacillus bataviensis (97.1 %), respectively. The cell wall peptidoglycan contained meso-diaminopimelic acid and the major isoprenoid quinone was MK-7. Major fatty acids are iso-C15:0 (32.4 %) and anteiso-C15:0 (27.4 %). Predominant polar lipids are diphosphatidylglycerol, phosphatidylglycerol and phosphatidylethanolamine. The results of physiological and biochemical tests allowed the genotypic and phenotypic distinctiveness of strain IITR-54T with its phylogenetic relatives and suggest that the strain IITR-54T should be recognized as a novel species, for which the name Bacillus mesophilum sp. nov. is proposed. The type strain is IITR-54T (= MTCC 11060T = JCM 19208T).

Similar content being viewed by others
References
Ash C, Farrow JAE, Wallbanks S, Collins MD (1991) Phylogenetic heterogeneity of the genus Bacillus as revealed by comparative analysis of small-subunit-ribosomal RNA sequences. Lett Appl Microbiol 3:202–206
Ash C, Priest FG, Collins MD (1993) Molecular identification of rRNA group 3 bacilli (Ash, Farrow, Wallbanks and Collins) using a PCR probe test. Proposal for the creation of a new genus Paenibacillus. Antonie Van Leeuwenhoek 64:253–260
Bergmann JG, Sainik J Jr (1957) Determination of trace amounts of chlorine in naptha. Anal Chem 29:241–243
Bligh EG, Dyer WJ (1959) A rapid method of total lipid extraction and purification. Can J Biochem Physiol 37:911–917
Boyle AF, Silvin CJ, Hassett JP, Nakas JP, Tanenbaum SW (1992) Bacterial PCB biodegradation. Biodegradation 3:285–298
Cowan ST, Steel KJ (1965) Manual for the identification of medical bacteria. Cambridge University Press, London
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Hatta T, Mukerjee-Dhar G, Damborsk J, Kiyohara H, Kimbara K (2003) Characterization of a novel thermostable Mn(II)-dependent 2,3-dihydroxybiphenyl 1,2-dioxygenase from a polychlorinated biphenyl- and naphthalene-degrading Bacillus sp. JF8. J Biol Chem 278:21483–21492
Kim O, Cho YJ, Lee K, Yoon SH, Kim M, Na H, Park SC, Jeon YS, Lee JH, Yi H, Won S, Chun J (2012) Introducing EzTaxon-e: a prokaryotic 16S rRNA Gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Microbiol 62:716–721
Kimbrough RD (1995) Polychlorinated biphenyls (PCBs) and human health: an update. Crit Rev Toxicol 25:133–163
Komagata K, Suzuki K (1987) Lipid and cell wall analysis in bacterial systematics. Methods Microbiol 19:161–206
Lanyi B (1987) Classical and rapid identification methods for medically important bacteria. Methods Microbiol 19:1–67
Mandel M, Marmur J (1968) Use of ultraviolet absorbance temperature profile for determining the gunine plus cytosine content of DNA. Methods Enzymol 12B:195–206
Manickam N, Misra R, Mayilraj S (2006) A novel pathway for the biodegradation of c-hexachlorocyclohexane by a Xanthomonas sp strain ICH12. J Appl Microbiol 102:1468–1478
Manickam N, Pathak A, Saini HS, Mayilraj S, Shanker R (2010) Metabolic profiles and phylogenetic diversity of microbial communities from chlorinated pesticides contaminated sites of different geographical habitats of India. J Appl Microbiol 109:1458–1468
Mayilraj S, Prasad GS, Suresh K, Saini HS, Shivaji S, Chakrabarti T (2005) Planococcus stackebrandtii sp. nov., isolated from a cold desert of the Himalayas, India. Int J Syst Evol Microbiol 55:91–94
Minnikin DE, O’Donnell AG, Goodfellow M, Alderson G, Athaly M, Schaal A, Parlett JH (1984) An integrated procedure for the extraction of isoprenoid quinones and polar lipids. J Microbiol Methods 2:233–241
Murray RG, Doetsch RN, Robinow CF (1994) Determinative and cytological light microscopy. In: Gerhard P, Murray RGE, Wood WA, Krieg NR (eds) Methods for general and molecular bacteriology. American Society for Microbiology, Washington, pp 21–41
Pieper DH, Reineke W (2000) Engineering bacteria for bioremediation. Curr Opin Biotechnol 11:262–270
Rainey FA, Stackebrandt E (1993) Phylogenetic evidence for the relationship of Saccharococcus thermophilus to Bacillus thermoglucosidasius, Bacillus kaustophilus and Bacillus stearothermophilus. Syst Appl Microbiol 16:224–226
Rainey FA, Fritze D, Stackebrandt E (1994) The phylogenetic diversity of thermophilic members of the genus Bacillus, as revealed by 16S rDNA analysis. FEMS Microbiol Lett 115:205–212
Sasser M (1990) Identification of bacteria by gas chromatography of cellular fatty acids. MIDI Technical note 101. MIDI Inc, Newark
Smibert RM, Krieg NR (1994) Phenotypic characterization. In: Gerhard P, Murray RGE, Wood WA, Krieg NR (eds) Methods for general and molecular bacteriology. American Society for Microbiology, Washington, pp 607–654
Smith NR, Gordon RE, Clark FE (1952) Aerobic spore forming bacteria. U.S. Department of Agriculture Monograph, no. 16
Stackebrandt E, Ebers J (2006) Taxonomic parameters revisited: tarnished gold standards. Microbiol Today 33:152–155
Stackebrandt E, Goebel BM (1994) Taxonomic note: a place for DNA–DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int J Syst Bacteriol 44:846–849
Staneck JL, Roberts GD (1974) Simplified approach to identification of aerobic actinomycetes by thin-layer chromatography. Appl Microbiol 28:226–231
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: evolutionary genetics analysis using maximum likelihood, evolutionary distance and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Tiedje JM, Quensen JF, Chee-Sanford J, Schimel JP, Boyd SA (1993) Microbial reductive dechlorination of PCBs. Biodegradation 4:231–240
Timmis KN, Steffan RJ, Unterman R (1994) Designing microorganisms for the treatment of toxic wastes. Annu Rev Microbiol 48:525–557
Wayne LG, Brenner DJ, Colwell RR et al (1987) International Committee on Systematic Bacteriology. Report of the adhoc committee on reconciliation of approaches to bacterial systematics. Int J Syst Bacteriol 37:463–464
Xu P, Li WJ, Tang SK, Zhang YQ, Chen GZ, Chen HH, Xu LH, Jiang CL (2005) Naxibacter alkalitolerans gen. nov., sp. nov., a novel member of the family ‘Oxalobacteraceae’ isolated from China. Int J Syst Evol Microbiol 55:1149–1153
Acknowledgments
We would like to thank Mr. Malkit Singh for his excellent technical assistance. This work was supported by CSIR (net work project NWP 006), and Government of India is duly acknowledged. This is IMTECH communication number 0132/2013.
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Erko Stackebrandt.
Natesan Manickam and Nitin Kumar Singh have contributed equally to the study.
CSIR-Institute of Microbial Technology (IMTECH) is a constituent laboratory of Council of Scientific and Industrial Research (CSIR), Government of India.
The GenBank accession number for the 16S rDNA sequence of strain IITR-54T is JN 210567.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Manickam, N., Singh, N.K., Bajaj, A. et al. Bacillus mesophilum sp. nov., strain IITR-54T, a novel 4-chlorobiphenyl dechlorinating bacterium. Arch Microbiol 196, 517–523 (2014). https://doi.org/10.1007/s00203-014-0988-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00203-014-0988-9