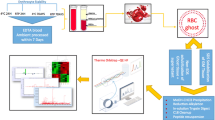
Abstract
Red blood cells (RBC) are the most common cell type found in blood. They might serve as reservoir for biomarker research as they are anuclear and lack the ability to synthesize proteins. Not many biomarker assays, however, have been conducted on RBC because of their large dynamic range of proteins, high abundance of lipids, and hemoglobin interferences. Here, we developed a semiquantitative mass spectrometry–based assay that targeted 144 proteins and compared the efficiency of urea, sodium deoxycholate, acetonitrile, and HemoVoid™ in their extraction of the RBC proteome. Our results indicate that protein extraction with HemoVoid™ led to hemoglobin reduction and increased detection of low abundance proteins. Although hemoglobin interference after deoxycholate and urea extraction was high, there were adequate amounts of low abundance proteins for quantitation. Extraction with acetonitrile led to an overall decrease in protein abundances probably as a result of precipitation. Overall, the best compromise in sensitivity and sample processing time was achieved with the urea–trypsin digestion protocol. This provided the basis for large-scale evaluations of protein targets as potential blood-based biomarkers. As a proof of concept, we applied this assay to determine that alpha-synuclein, a prominent marker in Parkinson’s disease, has an average concentration of approximately 40 μg mL−1 in RBC. This is important to know as the concentration of alpha-synuclein in plasma, typically in the picogram per milliliter range, might be partially derived from lysed RBC. Utilization of this assay will prove useful for future biomarker studies and provide a more complete analytical toolbox for the measurement of blood-derived proteins.

Graphical abstract





Similar content being viewed by others
Abbreviations
- AcN:
-
Acetonitrile
- AD:
-
Alzheimer’s disease
- ALS:
-
Amyotrophic lateral sclerosis
- APOE:
-
Apolipoprotein E
- CV:
-
Coefficient of variation
- DOC:
-
Sodium deoxycholate
- HV:
-
HemoVoid™
- LT:
-
LysC + Trypsin
- MRM:
-
Multiple reaction monitoring
- PD:
-
Parkinson’s disease
- PET:
-
Positron emission tomography
- RBC:
-
Red blood cell
- SRM:
-
Selected reaction monitoring
- T:
-
Trypsin
References
Lux SE. Anatomy of the red cell membrane skeleton: unanswered questions. Blood. 2016;127(2):187–99. https://doi.org/10.1182/blood-2014-12-512772.
Hünefeld FL. Der Chemismus in der thierischen Organisation. F A Brockhaus. 1840.
Weed RI, Reed CF, Berg G. Is hemoglobin an essential structural component of human erythrocyte membranes? J Clin Invest. 1963;42:581–8. https://doi.org/10.1172/JCI104747.
Wilmot VA, Swank RL. The influence of low-fat diet on blood lipid levels in health and in multiple sclerosis. Am J Med Sci. 1952;223(1):25–34.
Reed CF, Swisher SN, Marinetti GV, Enen EG. Studies of the lipids of the erythrocyte. I. Quantitative analysis of the lipids of normal human red blood cells. J Lab Clin Med. 1960;56:281–9.
Kakhniashvili DG, Bulla LA Jr, Goodman SR. The human erythrocyte proteome: analysis by ion trap mass spectrometry. Mol Cell Proteomics. 2004;3(5):501–9. https://doi.org/10.1074/mcp.M300132-MCP200.
Tyan YC, Jong SB, Liao JD, Liao PC, Yang MH, Liu CY, et al. Proteomic profiling of erythrocyte proteins by proteolytic digestion chip and identification using two-dimensional electrospray ionization tandem mass spectrometry. J Proteome Res. 2005;4(3):748–57. https://doi.org/10.1021/pr0497780.
Pasini EM, Kirkegaard M, Mortensen P, Lutz HU, Thomas AW, Mann M. In-depth analysis of the membrane and cytosolic proteome of red blood cells. Blood. 2006;108(3):791–801. https://doi.org/10.1182/blood-2005-11-007799.
Goodman SR, Daescu O, Kakhniashvili DG, Zivanic M. The proteomics and interactomics of human erythrocytes. Exp Biol Med (Maywood). 2013;238(5):509–18. https://doi.org/10.1177/1535370213488474.
Wilson MC, Trakarnsanga K, Heesom KJ, Cogan N, Green C, Toye AM, et al. Comparison of the proteome of adult and cord erythroid cells, and changes in the proteome following reticulocyte Maturation. Mol Cell Proteomics. 2016;15(6):1938–46. https://doi.org/10.1074/mcp.M115.057315.
Bryk AH, Wisniewski JR. Quantitative analysis of human red blood cell proteome. J Proteome Res. 2017;16(8):2752–61. https://doi.org/10.1021/acs.jproteome.7b00025.
Gautier EF, Leduc M, Cochet S, Bailly K, Lacombe C, Mohandas N, et al. Absolute proteome quantification of highly purified populations of circulating reticulocytes and mature erythrocytes. Blood Adv. 2018;2(20):2646–57. https://doi.org/10.1182/bloodadvances.2018023515.
Telen MJ. Erythrocyte adhesion receptors: blood group antigens and related molecules. Transfus Med Rev. 2005;19(1):32–44.
Kaul DK. Sickle red cell adhesion: many issues and some answers. Transfus Clin Biol. 2008;15(1–2):51–5. https://doi.org/10.1016/j.tracli.2008.03.012.
Iolascon A, Perrotta S, Stewart GW. Red blood cell membrane defects. Rev Clin Exp Hematol. 2003;7(1):22–56.
Mohandas N, Gallagher PG. Red cell membrane: past, present, and future. Blood. 2008;112(10):3939–48. https://doi.org/10.1182/blood-2008-07-161166.
Delaunay J. The molecular basis of hereditary red cell membrane disorders. Blood Rev. 2007;21(1):1–20. https://doi.org/10.1016/j.blre.2006.03.005.
Narla J, Mohandas N. Red cell membrane disorders. Int J Lab Hematol. 2017;39(Suppl 1):47–52. https://doi.org/10.1111/ijlh.12657.
de Lustig ES, Serra JA, Kohan S, Canziani GA, Famulari AL, Dominguez RO. Copper-zinc superoxide dismutase activity in red blood cells and serum in demented patients and in aging. J Neurol Sci. 1993;115(1):18–25.
Mohanty JG, Shukla HD, Williamson JD, Launer LJ, Saxena S, Rifkind JM. Alterations in the red blood cell membrane proteome in Alzheimer's subjects reflect disease-related changes and provide insight into altered cell morphology. Proteome Sci. 2010;8:11. https://doi.org/10.1186/1477-5956-8-11.
Varady G, Szabo E, Feher A, Nemeth A, Zambo B, Pakaski M, et al. Alterations of membrane protein expression in red blood cells of Alzheimer's disease patients. Alzheimers Dement (Amst). 2015;1(3):334–8. https://doi.org/10.1016/j.dadm.2015.06.007.
Zuckerman SH, Uretsky SC, Douglas SD. Partial purification of ABO blood group antigens: sodium deoxycholate fractionation of human erythrocyte membranes. Vox Sang. 1974;27(6):504–14.
Leon IR, Schwammle V, Jensen ON, Sprenger RR. Quantitative assessment of in-solution digestion efficiency identifies optimal protocols for unbiased protein analysis. Mol Cell Proteomics. 2013;12(10):2992–3005. https://doi.org/10.1074/mcp.M112.025585.
Moore SM, Hess SM, Jorgenson JW. Extraction, enrichment, solubilization, and digestion techniques for membrane proteomics. J Proteome Res. 2016;15(4):1243–52. https://doi.org/10.1021/acs.jproteome.5b01122.
Coleman R, Holdsworth G, Finean JB. Detergent extraction of erythrocyte ghosts. Comparison of residues after cholate and Triton X-100 treatments. Biochim Biophys Acta. 1976;436(1):38–44.
Barasa B, Slijper M. Challenges for red blood cell biomarker discovery through proteomics. Biochim Biophys Acta. 2014;1844(5):1003–10. https://doi.org/10.1016/j.bbapap.2013.10.002.
Walpurgis K, Kohler M, Thomas A, Wenzel F, Geyer H, Schanzer W, et al. Validated hemoglobin-depletion approach for red blood cell lysate proteome analysis by means of 2D PAGE and Orbitrap MS. Electrophoresis. 2012;33(16):2537–45. https://doi.org/10.1002/elps.201200151.
Marx V. Targeted proteomics. Nat Methods. 2013;10(1):19–22.
Bartels T, Choi JG, Selkoe DJ. alpha-Synuclein occurs physiologically as a helically folded tetramer that resists aggregation. Nature. 2011;477(7362):107–10. https://doi.org/10.1038/nature10324.
Fauvet B, Mbefo MK, Fares MB, Desobry C, Michael S, Ardah MT, et al. alpha-Synuclein in central nervous system and from erythrocytes, mammalian cells, and Escherichia coli exists predominantly as disordered monomer. J Biol Chem. 2012;287(19):15345–64. https://doi.org/10.1074/jbc.M111.318949.
Lothian A, Lago L, Mukherjee S, Connor AR, Fowler C, McLean CA, et al. Characterization of the metal status of natively purified alpha-synuclein from human blood, brain tissue, or recombinant sources using size exclusion ICP-MS reveals no significant binding of Cu. Fe or Zn Metallomics. 2018. https://doi.org/10.1039/c8mt00223a.
Roberts BR, Doecke JD, Rembach A, Yevenes LF, Fowler CJ, McLean CA, et al. Rubidium and potassium levels are altered in Alzheimer's disease brain and blood but not in cerebrospinal fluid. Acta Neuropathol Commun. 2016;4(1):119. https://doi.org/10.1186/s40478-016-0390-8.
Ellis KA, Bush AI, Darby D, De Fazio D, Foster J, Hudson P, et al. The Australian Imaging, Biomarkers and Lifestyle (AIBL) study of aging: methodology and baseline characteristics of 1112 individuals recruited for a longitudinal study of Alzheimer's disease. Int Psychogeriatr. 2009;21(4):672–87. https://doi.org/10.1017/S1041610209009405.
Leong SL, Hinds MG, Connor AR, Smith DP, Illes-Toth E, Pham CL, et al. The N-terminal residues 43 to 60 form the interface for dopamine mediated alpha-synuclein dimerisation. PLoS One. 2015;10(2):e0116497. https://doi.org/10.1371/journal.pone.0116497.
Proc JL, Kuzyk MA, Hardie DB, Yang J, Smith DS, Jackson AM, et al. A quantitative study of the effects of chaotropic agents, surfactants, and solvents on the digestion efficiency of human plasma proteins by trypsin. J Proteome Res. 2010;9(10):5422–37. https://doi.org/10.1021/pr100656u.
Zhou J, Zhou T, Cao R, Liu Z, Shen J, Chen P, et al. Evaluation of the application of sodium deoxycholate to proteomic analysis of rat hippocampal plasma membrane. J Proteome Res. 2006;5(10):2547–53. https://doi.org/10.1021/pr060112a.
MacLean B, Tomazela DM, Shulman N, Chambers M, Finney GL, Frewen B, et al. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010;26(7):966–8. https://doi.org/10.1093/bioinformatics/btq054.
Chong J, Soufan O, Li C, Caraus I, Li S, Bourque G, et al. MetaboAnalyst 4.0: towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 2018;46(W1):W486–W94. https://doi.org/10.1093/nar/gky310.
Kiko T, Nakagawa K, Satoh A, Tsuduki T, Furukawa K, Arai H, et al. Amyloid beta levels in human red blood cells. PLoS One. 2012;7(11):e49620. https://doi.org/10.1371/journal.pone.0049620.
Baillet A, Chanteperdrix V, Trocme C, Casez P, Garrel C, Besson G. The role of oxidative stress in amyotrophic lateral sclerosis and Parkinson's disease. Neurochem Res. 2010;35(10):1530–7. https://doi.org/10.1007/s11064-010-0212-5.
Matsumoto Y, Murakami H, Hattori N, Yoshimoto K, Asano S, Inden M. Excessive expression of hippocampal ezrin is induced by intrastriatal injection of 6-hydroxydopamine. Biol Pharm Bull. 2011;34(11):1753–8.
Johannsen P, Velander G, Mai J, Thorling EB, Dupont E. Glutathione peroxidase in early and advanced Parkinson's disease. J Neurol Neurosurg Psychiatry. 1991;54(8):679–82.
Lin CH, Yang SY, Horng HE, Yang CC, Chieh JJ, Chen HH, et al. Plasma alpha-synuclein predicts cognitive decline in Parkinson's disease. J Neurol Neurosurg Psychiatry. 2017;88(10):818–24. https://doi.org/10.1136/jnnp-2016-314857.
Bougea A, Stefanis L, Paraskevas GP, Emmanouilidou E, Vekrelis K, Kapaki E. Plasma alpha-synuclein levels in patients with Parkinson's disease: a systematic review and meta-analysis. Neurol Sci. 2019;40(5):929–38. https://doi.org/10.1007/s10072-019-03738-1.
Vicente Miranda H, Cassio R, Correia-Guedes L, Gomes MA, Chegao A, Miranda E, et al. Posttranslational modifications of blood-derived alpha-synuclein as biochemical markers for Parkinson's disease. Sci Rep. 2017;7(1):13713. https://doi.org/10.1038/s41598-017-14175-5.
Percy AJ, Chambers AG, Smith DS, Borchers CH. Standardized protocols for quality control of MRM-based plasma proteomic workflows. J Proteome Res. 2013;12(1):222–33. https://doi.org/10.1021/pr300893w.
Percy AJ, Yang J, Chambers AG, Simon R, Hardie DB, Borchers CH. Multiplexed MRM with internal standards for cerebrospinal fluid candidate protein biomarker quantitation. J Proteome Res. 2014. https://doi.org/10.1021/pr500317d.
Yeung YG, Stanley ER. Rapid detergent removal from peptide samples with ethyl acetate for mass spectrometry analysis. Curr Protoc Protein Sci. 2010;16(16):2. https://doi.org/10.1002/0471140864.ps1612s59.
Tu C, Rudnick PA, Martinez MY, Cheek KL, Stein SE, Slebos RJ, et al. Depletion of abundant plasma proteins and limitations of plasma proteomics. J Proteome Res. 2010;9(10):4982–91. https://doi.org/10.1021/pr100646w.
Acknowledgments
We would like to acknowledge the research team of The Australian Imaging, Biomarker & Lifestyle Flagship Study of Ageing (AIBL; a complete list of researchers can be found at www.aibl.csiro.au) and the volunteers and their families. We would also like to thank the Victorian Government’s Operational Infrastructure Support Program, the Wicking Trust and The Florey Institute of Neuroscience and Mental Health Neuroproteomics Facility. Furthermore, we would like to thank the Melbourne Mass Spectrometry and Proteomics Facility of The Bio21 Molecular Science and Biotechnology Institute (University of Melbourne) for the support of mass spectrometry analysis. We also thank Alicia Siew for assistance in testing protocols used for the development of these assays and Oliver R. B. Thomas for his comments and critical review of the manuscript.
Funding
We acknowledge funding and support; BR receives partial support from the National Health and Medical Research Council (1061550 & 1138673), and Motor Neuron Disease Research Institute of Australia and the Neuroproteomics Facility. BR and SK receive support from the Michael J. Fox Foundation (MJFF, Parkinson’s Research). BR receives support from Alzheimer’s Disease Drug Discovery Foundation. BR receives research support from Agilent Technologies and e-MSion, Inc.
Author information
Authors and Affiliations
Contributions
S. Klatt designed the research, performed the experiments, optimized the mass spectrometry-based assay, analyzed the results and wrote the paper. A. Roberts and A. Lothian co-developed RBC protein extraction and mass spectrometry-based assay. C. Masters and C. Fowler identified blood samples from the AIBL study and were involved in the experimental design. R. Cappai designed the alpha-synuclein plasmid and designed the expression and purification protocols. B. Roberts designed the research, co-developed the mass spectrometry-based assay, analyzed the results and wrote the paper.
Corresponding author
Ethics declarations
Conflict of interest disclosure
BR receives research support from Agilent Technologies, Biosensis and eMSion. All other authors declare to have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(PDF 5246 kb)
Rights and permissions
About this article
Cite this article
Klatt, S., Roberts, A., Lothian, A. et al. Optimizing red blood cell protein extraction for biomarker quantitation with mass spectrometry. Anal Bioanal Chem 412, 1879–1892 (2020). https://doi.org/10.1007/s00216-020-02439-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-020-02439-5