Abstract
As a more efficient and effective way to address disease diagnosis and intervention, cutting-edge technologies, devices, therapeutic approaches, and practices have emerged within the personalized medicine concept depending on the particular patient's biology and the molecular basis of the disease. Personalized medicine is expected to play a pivotal role in assessing disease risk or predicting response to treatment, understanding a person's health status, and, therefore, health care decision-making. This work discusses electrochemical biosensors for monitoring multiparametric biomarkers at different molecular levels and their potential to elucidate the health status of an individual in a personalized manner. In particular, and as an illustration, we discuss several aspects of the infection produced by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) as a current health care concern worldwide. This includes SARS-CoV-2 structure, mechanism of infection, biomarkers, and electrochemical biosensors most commonly explored for diagnostics, prognostics, and potentially assessing the risk of complications in patients in the context of personalized medicine. Finally, some concluding remarks and perspectives hint at the use of electrochemical biosensors in the frame of other cutting-edge converging/emerging technologies toward the inauguration of a new paradigm of personalized medicine.
Graphical abstract

Similar content being viewed by others
Introduction
Personalized medicine is a group of cutting-edge technologies, devices, interventions, and practices designed on demand based on the characteristics of the individual, to be used for decision-making to determine risk of disease or predict response to treatment [1]. Personalized medicine is also called precision, stratified, or P4 medicine, often used interchangeably [2, 3]. Notably, it refers to new transformative diagnostic and informatics approaches coupled with an understanding of the disease's molecular basis for patient stratification. Appropriately specific diagnostic and therapeutic parameters are selected based on the patient's fundamental systems biology and dynamics, i.e., DNA, RNA expression, translation, and levels of proteins [4,5,6]. Therefore, it may include molecular or cellular analysis by genomics and proteomics [7], medical imaging, nanoparticle-based theranostics (therapeutics and diagnostics) [8], or toxgnostics (personalized drug toxicity), among others, resulting in a broader understanding of a person's health status. Furthermore, combining clinical data and risk factors with genomics, proteomics, and imaging provides a diagnosis of pathology and information about the prognosis, prediction, and recurrence of a disease, and susceptibility and survivability of the patient [9]. Therefore, detecting and monitoring biomarkers of different molecular levels is very effective, especially when screening populations to identify disease risks and implement timely preventive efforts [10].
Personalized medicine promises to play a pivotal role in clinical practice as a prospective approach to making optimal individual health care decisions. It uses predictive tools to evaluate health risks and design personalized health plans to help patients mitigate risks, prevent disease, and treat it with precision when it occurs. Personalized health care can be tailored based on the patient's genetic makeup, as mentioned, or on the disease-causing agent, such as drug-resistant bacteria or viruses [11]. In this context, personalized medicine may permit the customization of a unique treatment approach specific to the individual biology and genome, providing better diagnoses and earlier treatment, more efficient drug development, and targeted therapeutics. Looking at a patient as an individual enables a comparison with other healthy or ill individuals to detect differences that account for potential diseases, thereby facilitating a more accurate diagnosis and implementation of a specific treatment plan and prevention of adverse events.
One of the most critical aspects guiding therapy is early and proper diagnostics. Nowadays, it involves genomics, in vivo imaging with contrast and fluorescent agents or nanoparticle-based markers, nuclear imaging agents [12], and in vitro laboratory tests [8] that often combine molecular assays [13] with deep learning algorithms and artificial intelligence in search of disease-specific biomarkers [14]. Along with diagnostics, such approaches hold great promise for preventive care. In this context, companion diagnostics [15] are used with a therapeutic drug to determine its potential applicability to the individual.
Once a disease is diagnosed, a more proper alternative for finding a treatment can be pursued which, unlike trial and error, can realize "therapy with the right drug at the right dose in the right patient" [16], promoted by personalized medicine. The potential treatment can be tailored based on how the patient would respond considering its genome and, therefore, can be more efficacious, accurate, and cost-effective. Besides, the customized production of a drug, varying ingredients, dose level, or administration route, among other features, supported by computational and mathematical models to study drug interactions, pharmacodynamics, and pharmacokinetics, is accepted within the concept of personalized medicine, on the path to replace mass-produced doses or fixed-dose combinations. In the same context, the development of functional nanocarriers [17, 18] for site-specific and controlled drug delivery is a branch of personalized medicine that promises to revolutionize health care. Finally, theranostics refers to a personalized approach to treating a pathology, especially cancer, with similar molecules or approaches for diagnosis and therapy.
The following sections discuss the potential of electrochemical biosensors for the multiparametric monitoring of biomarkers at different molecular levels and how they can contribute to advancing more efficient health care strategies. As a novel and illustrative example, we selected the infection produced by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), a global concern and impact pathology, to exemplify the role of electrochemical biosensors in assessing patients' diagnostics and risk of complications in the frame of personalized medicine approaches. First, we introduce general aspects of SARS-CoV-2, including the structure, infection mechanism [19, 20], and signature of biomarkers for diagnosis, prognosis, and prediction of the course of the disease. Then, we review electrochemical biosensors for SARS-CoV-2 detection [21,22,23], prognosis, and prediction, to end with some concluding remarks and perspectives toward a new paradigm of personalized medicine.
SARS-CoV-2 general aspects
Although zoonotic pathogens (animal origin) have affected humanity since the beginning of the twenty-first century [24,25,26,27], the recent emergence of viral infectious diseases caused by coronaviruses has affected the entire global population. Coronaviruses are a group of single-stranded RNA viruses named for the corona-like structure on their outer surface, classified into alpha (α), beta (β), gamma (γ), and delta (δ) genera. Although these viruses infect a wide variety of mammalian (alpha and beta) and avian (gamma and delta) species, only seven are known to have infected humans [25, 28,29,30]. Four of these coronaviruses have low pathogenicity, being endemic in humans, namely, human coronavirus OC43 (HCoV-OC43), HKU1 (HCoV-HKU1), NL63 (HCoV-NL63), and 229E (HCoV-229E), but the other three, i.e., severe acute respiratory syndrome coronavirus (SARS-CoV), Middle East respiratory syndrome coronavirus (MERS-CoV), and SARS-CoV-2, are highly pathogenic [25, 29, 31]. This last-mentioned is indeed a beta coronavirus responsible for severe and potentially fatal respiratory disease [25, 29].
SARS-CoV, transmitted from civet cats to humans, emerged in China in 2002 and caused a pandemic, with more than 8000 infected and 800 deaths. MERS-CoV, transmitted from dromedaries to humans, emerged in 2012 in the Middle East, causing about 2500 cases and 860 deaths worldwide [25, 32]. In December 2019, in Wuhan, Hubei province of China, a new coronavirus, SARS-CoV-2, was found to be responsible for an outbreak of atypical pneumonia defined as coronavirus disease 2019 (COVID-19) [31, 32]. In January 2020, the virus was sequenced and isolated in China [31], and since then it has spread rapidly worldwide. The World Health Organization (WHO) declared it a pandemic on March 11, 2020 [29], and about 2 years later we had more than 400 million cases and 5.76 million deaths globally [33], even though by February 2022, 54% of people were already fully vaccinated [34].
Structure of SARS-CoV-2
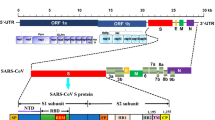
SARS-CoV-2 shares approximately 80% of its genome with SARS-CoV and 96% with the bat coronavirus BatCoV RaTG13 [29, 30], indicating that SARS-CoV-2 most likely originated in bats and jumped to humans via an intermediary that research seems to indicate was the pangolin [25]. Its RNA sequence is approximately 30,000 bases long and its structure consists of four proteins, i.e., spicule (S), membrane (M), envelope (E), and nucleocapsid (N) proteins (see Fig. 1) [36].
Structure of SARS-CoV-2, formed mainly by the genetic material (RNA) and the four structural proteins, i.e., N, E, M, and S proteins. Modified from reference [35]
The SARS-CoV-2 genome contains two 5′ and 3′ terminal end regions (UTRs), one open reading frame (ORF) encoding the 16 nonstructural proteins (nsp1–16), and five other ORFs encoding the four structural proteins and eight accessory proteins [29]. The highly immunogenic phosphoprotein N protein is the most abundant in the coronavirus [37]. The N protein consists of an N-terminal domain and a C-terminal domain, both of which can bind to the viral RNA and thus help with the packaging of the viral genome [36] responsible for viral RNA transcription and replication [38]. The E protein is an integral membrane protein of 8–12 KDa and regulates virus life cycles, such as virus assembly and release. In addition, it functions as an ion channel, which is necessary for pathogenesis [29, 36]. The M protein is a dimeric protein with a size of 25–30 KDa, which is responsible for shaping the virion, as it maintains the curvature of the membrane and binds to the nucleocapsid [36]. The binding of M and N proteins stabilizes the nucleocapsid and helps complete viral assembly, while the E and M proteins form the viral envelope [39]. The S protein is a 150-KDa trimeric class I fusion glycoprotein located on the viral particle surface responsible for binding to the host receptor. It is highly N-glycosylated and forms peaks between 18 and 23 nm long on the virus surface. It consists of S1 and S2 subunits [36, 40]. The S1 subunit is responsible for receptor recognition. It comprises four distinct A–D domains, consisting of the N-terminal region (NTD) formed by the A domain, also called the receptor-binding domain (RBD), and the C-terminal region (CTD) formed by B, C, and D domains. The S2 subunit facilitates virus–cell fusion and forms the stem of the spike [25, 29, 39, 40].
Understanding the SARS-CoV-2 mechanism of infection
SARS-CoV-2 is transmitted from person to person by direct inhalation of contaminated droplets released into the environment when an infected person sneezes or coughs. It is also transmitted by direct contact through oral, nasal, and ocular mucosa. Other important means of transmission are objects that have been in contact with an infected person [35, 41]. Upon entering the body, SARS-CoV-2 binds to epithelial cells in the mouth or nose and can even migrate through the airways and infect type II alveolar pneumocytes [35]. These cells are characterized by having within their receptors the human angiotensin-converting enzyme 2 (ACE2), being the main entry point of the virus, even though other SARS-CoV-2 entry receptors have also been reported, for example, DC-SIGN (CD209), CD147, L-SIGN (CD209L) [41], and AXL [42]. ACE2 is found in various tissues and organs, including the lungs, heart, kidneys, liver, gastrointestinal tract, and blood vessels. This enzyme regulates blood pressure and inflammation by regulating the renin–angiotensin–aldosterone system [25, 41].
The RBD in the S protein from the virus consists of 394 glutamine residues, recognized explicitly by 31 lysine residues of the host ACE2 enzyme [35, 43]. The RBD region of the SARS-CoV-2 binds to the ACE2 receptor with 10–20-fold higher affinity than SARS-CoV, which facilitates viral entry and explains the ease of virus spread from person to person [43, 44]. Following the binding between the S protein and the ACE2 receptor, the acid-dependent transmembrane protease serine 2 (TMPRSS2), cathepsins, and furin enzymes carry out the S protein cleavage precisely in two portions of the S2 region. In the first one, the RBD region and the fusion domains of the S protein are separated, and in the second one, the fusion peptide is exposed and inserted into the membrane to allow the fusion of the viral and host membranes [36, 39, 45,46,47]. When the virus enters the cell, RNA is released into the cell cytoplasm and initiates its translation and replication process by appropriating its reproductive machinery (endoplasmic reticulum and Golgi complex and the endoplasmic reticulum) to produce more viral copies. Finally, the virus is transported to the membrane, exits the cell by exocytosis, and travels to infect other cells [25, 35, 39].
COVID-19 biomarkers: from diagnosis to prognosis and prediction of the course of the disease
Biomarkers are biomolecules that act as biological indicators of the presence, severity, or type of a disease. They play a fundamental role in diagnosis and prediction of disease severity and even future complications [25]. In particular in COVID-19 management, apart from diagnosis, some biomarkers have been associated with evaluating disease progression, as summarized in Fig. 2 and Table 1.
Overview of the present and future of COVID-19-associated biomarker detection. Briefly, precision medicine of COVID-19 can be achieved by (a) diagnosing infection from the detection of genetic material, structural proteins, and IgG and IgM antibodies, and (b) detecting inflammatory [C-reactive protein (CrP), interleukin 6 (IL-6), procalcitonin (PCT), ferritin (FT)], hematological [lymphocyte count (L), neutrophil count (N), N/L ratio (NLR)], and biochemical [D-dimer, cardiac troponin (cTn), creatine kinase (CK), vitamin D] biomarkers that predict disease prognosis, progression, and severity. In addition, moving from conventional detection to using (c) different robust commercial detection kits requiring specialized equipment and personnel to (d) a single multiparametric device based on sensitive, specific and portable nanobiosensors [48, 49]
Biomarkers for diagnosis
Although personalized medicine has been commonly exploited in cancer research, it has found step-by-step applications in various pathologies, such as respiratory diseases. Respiratory diseases affect the population worldwide, with particular mortality in low-income countries. However, respiratory diseases are heterogeneous and often share symptoms, complicating and delaying diagnosis. In this context, personalized medicine offers invaluable diagnostic alternatives based on proteomics in which, unlike a single biomarker, a series of protein expression and a diversity of body fluids can be analyzed [101]. Pathogen detection aims to identify specific biomolecules of the microorganism or molecular changes in the host [25]. Currently, there are three strategies to detect SARS-CoV-2, namely, detection of the viral genetic material (RNA), viral antigens (structural proteins), or antibodies generated by the host (see Fig. 2a) [102].
The main diagnostic tool for SARS-CoV-2 has been reverse transcription polymerase chain reaction (RT-PCR) (see Table 1), a technique that detects the genetic material of the virus by combining reverse transcription of RNA into complementary DNA and amplification of specific targets [25, 103, 104]. The sample is initially collected from the patient mainly by nasopharyngeal swab, followed by RNA extraction and purification for reverse transcription. Finally, the sample reacts with a cocktail of probes that recognize specific SARS-CoV-2 biomarkers such as the E gene, the RdRp gene, and the ORF 1ab gene in a thermal cycler [105,106,107]. Because of its high sensitivity and specificity, this technique is the most widely used for accurate and reliable identification of the virus [29]. However, it requires highly specialized personnel and instrumentation and between 2 and 5 hours to obtain the results; moreover, due to the need to transport the samples to specialized laboratories, it can take 24 hours or more, increasing the related costs and the possibility of viral spread [38, 107].
Another strategy currently used to detect SARS-CoV-2 is rapid antigen detection tests (RADTs) (see Table 1) [108], which detect viral particles from their structural proteins such as S protein or N protein. Although less sensitive than RT-PCR, it is faster and easier to implement, obtaining results in approximately 30 minutes [108, 109]. In addition, the detection of genetic material and structural proteins is directly related to the viral load, considered a predictor of the severity and progression of the disease, presenting the most critical states and higher probability of death in patients with higher viral load [110].
Serological tests have also been used to detect SARS-CoV-2 infection, an indirect method that detects antibodies generated by the body of infected people. When foreign agents such as microorganisms enter the body, the immune system responds rapidly to eliminate the foreign agents and creates antibodies that detect them to afford some future immunity [111]. In the case of COVID-19, the body generates immunoglobulin M (IgM) present at the beginning of the infection or when it is acute, and immunoglobulin G (IgG) appears as a response to the acute phase. For their detection, a blood sample is taken from the patient and transferred directly to the test (immunochromatographic assay, enzyme-linked immunosorbent assay [ELISA], or lateral flow immunoassay) (see Table 1), which gives the results in 5–15 minutes up to hours [112,113,114]. However, these tests are not recommended as diagnostic tools due to their low sensitivity. For example, in asymptomatic cases, the concentration of these antibodies is low, so false-negative results can be obtained in people who have the infection [114]. In addition, a positive result only indicates that the person has been in contact with the virus, and it is not known precisely whether the person is still infected when the sample is taken [115].
Biomarkers for prognosis and prediction of the course of the disease
COVID-19 patients may be asymptomatic or, in many cases, present with pneumonia, acute respiratory distress syndrome (ARDS), multi-organ failure, and, in some cases, even death. Different pathways of clinical manifestation of COVID-19 have been reported, including a high inflammatory response, low white blood cell and lymphocyte counts, and abnormal coagulation parameters. These complications are associated with proteins and genetic factors expressed differently in each person and are related to the progression and severity of the disease (see Fig. 2b) [116]. The following are some of the most relevant biomarkers in a person's susceptibility to severe symptoms and even death, summarized in Table 1, showing the conventional and point-of-care (POC) detection strategies available on the market.
Inflammatory biomarkers
Experience from studies in SARS-CoV and MERS-CoV has demonstrated a relationship between the inflammatory response called cytokine storm produced in response to these viruses and the progression of these viral infections [117,118,119,120,121]. Excessive release of inflammatory biomarkers has also been observed in COVID-19 [122] in response to the infection [123], thus triggering viral sepsis and lung injury and leading to various complications and eventually a fatal outcome [124]. Furthermore, different studies have shown changes in the levels of inflammatory cytokines such as interleukin 1 (IL-1), interleukin 6 (IL-6), interleukin 8 (IL-8), interleukin 10 (IL-10), and tumor necrosis factor alpha (TNF-α) and chemokines such as C-X-C motif chemokine ligand 10 (CXCL10), chemokine ligand 3 (CCL3), and monocyte chemoattractant protein 1 (MCP1) in patients with COVID-19 [125, 126]. High levels of IL-6, IL-10, TNF-α have even been reported in patients with post-acute sequelae of infection three months after diagnosis [127].
IL-6 is a glycoprotein involved in various immunomodulatory and inflammatory processes and is one of the main biomarkers of COVID-19 severity, produced due to tissue injury and infection [128], and assists in the maturation of B cells [129]. In addition, IL-6 is involved in the acute phase and chronic inflammation [130] and is a biomarker of sepsis. While the normal range of IL-6 is from 0 to 16.4 pg/ml [131], levels in patients with complicated COVID-19 are three times as high as those with uncomplicated disease [132]. Furthermore, this biomarker is critical in the cytokine storm, as it activates several cells and other acute-phase biomarkers such as C-reactive protein [133].
Other widely studied inflammatory biomarkers include C-reactive protein (CrP), procalcitonin (PCT) and ferritin (FT). CrP is a biomarker that may be present in the bloodstream at the time of infection, produced by liver cells in response to inflammation [126, 134] . While CrP levels below 0.3 mg/dL are considered normal in healthy adults [135], concentrations in patients with COVID are higher. High CrP levels of 20–50 mg/L may be early indicators of how COVID-19 will progress [135]. Therefore, this biomarker becomes a "fallback" in the case of inflammation, commonly observed in SARS-CoV-2 infections. PCT is a precursor of the hormone calcitonin, expressed in cells due to infection or injury. It is a possible diagnostic biomarker of sepsis, and a level of PCT below 0.15 ng/ml is considered normal [136]. Above this level, the risk of severe infection is more than five times that in patients with low PCT values [133, 137]. It has been reported as a sepsis biomarker more specific than IL-6 and other biomarkers for early detection [138, 139]. Ferritin (FT) is an iron storage protein essential in regulating the body's iron content and oxidative stress and anemia, and more recently it has been associated with SARS-CoV-2 viral infections. According to WHO, normal ferritin levels should range from 15 to 200 ng/mL and 15 to 150 ng/mL for men and women, respectively. However, patients with severe COVID-19 have shown ferritin levels above 1000 ng/mL [126, 140].
A new and lesser-known biomarker is LIGHT, a cytokine encoded by the tumor necrosis factor superfamily member 14 (TNFSF14) gene that plays a crucial role in regulating the immune response. LIGHT is homologous to lymphotoxin, exhibits inducible expression, and competes with herpes simplex virus (HSV) glycoprotein D for binding to herpesvirus entry mediator (HVEM), a receptor expressed on T lymphocytes [141]. Elevated cytokine levels stimulate T-cell and B-cell responses, inducing the release of other cytokines such as IL-1, IL-6, IL-8, IL-10, TNF, and granulocyte-macrophage colony-stimulating factor (GM-CSF), and driving the cytokine storm [142]. For example, levels above 250 pg/mL were reported in persons with acute lung injury pneumonia associated with SARS-CoV-2 infection [143].
Hematological biomarkers
COVID-19 manifests with hematological alterations such as leukocytosis, leukopenia, lymphocytopenia, eosinopenia, neutrophilia, and thrombocytopenia [144]. Low white blood cell and lymphocyte counts are related to disease stage (between days 7 and 14 after infection) and severity. While lymphocyte counts below 1100 cells/μL have been reported in patients with severe disease and non-survivors [133, 145], it has been reported that the leukocyte and lymphocyte count is within average levels in asymptomatic patients. It has also been demonstrated that the virus causes lymphocyte lysis, since these cells express ACE2 receptors. During the virus incubation time, the individual presents an average lymphocyte count, but after one or two weeks, the atrophy of the lymphoid organs decreases their production. In addition, high lactate dehydrogenase (LDH) concentrations inhibit the proliferation of lymphocytes [133, 144]. A significant decrease mainly in T lymphocytes, especially TCD4 and TCD8 cells, leads to the development of lymphopenia in patients with severe disease [125]. This decrease occurs during the first week after infection and gradually increases after the second week [125], which generates lymphocytopenia, directly related to the severity of COVID-19 [133, 145]. It has also been found that T-cell counts correlate inversely with blood cytokine levels in patients with severe disease.
Neutrophils have also been shown to increase [133, 145], which, together with the low levels of lymphocytes, modifies the neutrophil–lymphocyte ratio (NLR). The NLR and the platelet–lymphocyte ratio (PLR) were shown to be potential biomarkers for predicting mortality and prognosis of the disease. Average NLR values below three have been reported, and values above this level have been reported to indicate infection, with a high probability of sepsis when values increase to above nine [144].
Endothelial biomarkers
Endothelial cells are responsible for maintaining the integrity of the vascular endothelium and inhibiting excessive clotting [126]. An increase in endothelial proteins and platelet activation molecules has been evidenced in patients with severe COVID-19. Endothelin 1 (ET-1) is a vasoconstrictor that increases its secretion in the presence of angiotensin II, cytokines, and free radicals, characteristic in severe cases of the disease. Another possible endothelial biomarker is syndecan 1, a transmembrane protein that indicates vascular endothelial activation and inhibits epithelial wound healing in the alveoli, promoting pulmonary fibrosis [146]. Elevated levels of this biomarker have been reported in patients with severe COVID-19 (336.5 ng/mL) relative to healthy patients (41.5 ng/mL) [126].
Biochemical biomarkers
Biochemical biomarkers including D-dimer, cardiac troponin (cTn), and LDH have become of great interest in the progression and prognosis of COVID-19, because they are widely associated with increased susceptibility to severe disease and a high risk of mortality. D-dimer originates from the lysis of cross-linked fibrin and is an indicator of coagulation and fibrinolysis [126, 147]. Therefore, D-dimer is related to disease severity and might be an early disease biomarker. While values lower than 0.5 μg/mL were associated with hospitalized patients without intensive care unit (ICU) admission [148], levels above 2.0 μg/mL were predictive of mortality [126, 133], as these elevated levels indicated a state of hypercoagulability in patients [149], which is systemic and can lead to limb ischemia and even coagulation factor deterioration [147]. cTn is a protein related to myocardial lesions [150], and this, in turn, with a tripling of the risk of mortality [151]. Increased biomarker concentration has been associated with disease severity [152], with a higher probability of death at concentrations above 28 ng/L [150]. High concentrations of cTn (> 0.03 ng/mL) were associated with elevated concentrations of D-dimer, CrP, LDH, and PCT [151, 152]. LDH is an indicator of acute tissue damage to the heart, liver, lungs, muscles, and kidneys [153]. Abnormal LDH values are related to decreased oxygenation, leading to multiple organ damage. In addition, it has been reported that elevated values are associated with a sixfold and 16-fold increased probability of severe disease and mortality, respectively, with a cutoff value of 263.5 U/L [110]. Therefore, this biomarker could be used to predict the severity of COVID-19 [154].
Other biomarkers
Other biomarkers have been associated with severe cases of COVID-19 but are less often reported than those described above. However, they could provide a more personalized approach to predicting and treating the disease. These include creatine kinase (CK), bradykinin (BK), microRNAs, testosterone, and even the pulmonary microbiota. CK is an enzyme in different body tissues that acts as a biomarker of muscle damage [154, 155]. An increase in this biomarker above 200 U/L has been reported in severe cases of COVID-19, related mainly to muscle pain, respiratory failure, and a high risk of death [155,156,157], but it is less common than other biomarkers, reporting a sensitivity of only 28% [156]. BK is a peptide responsible for regulating blood pressure, and its levels are increased with increased ACE2, generating a bradykinin storm, which can generate characteristic manifestations of COVID-19 such as ARDS, inflammation, and edema [126]. In addition, it has been reported to cause pain, vessel expansion, and even endothelial dysfunction [126].
MicroRNAs are non-coding RNAs that regulate post-transcriptional expression in various processes such as cell proliferation, apoptosis, and differentiation. It has been reported that they can alter the expression of ACE2 and TMPRSS2 during COVID-19 [126] and modulate the immune response to disease. Some of those studied include miR-376a-3p, miR-99b-5p, miR-10a-5p, miR-376a-3p, miR-548av-5p, and miR-99b-5p [126], in addition to miR-21-5p, miR-146a, miR-126-3p, miR-144, and miR-155; the circulating miR-21-5p, miR-144, and miR-155 are mainly related to disease diagnosis and progression [158]. Genes can be up- and downregulated in patients with diseases such as COVID-19, where about 2289 upregulated genes and 912 downregulated genes have been reported in the presence of the disease, most of which are part of the immune response [159]. Overexpression of genes such as CD177, S100A12, ELANE, OLFM4, MPO, RETN, ARG1, CD15, S100A8/9, PADI4, NLRC4, MMP8 and MMP9, PRDM1, XBP1, and IRF4, and downregulation of genes such as CX3CR1 and MSR1 TRAC, TRBC1, CD247, CD4, CD2, TBET, and IL7R are associated with disease severity [110].
The concentrations of total testosterone (TT) and calculated free testosterone (cFT) have also been used as biomarkers of severity, showing a progressive decrease with disease progression and a higher risk of ICU hospitalization and death in men with TT < 5 nmol/L or cFT < 100 pmol/L. These data corroborate the disease trends, where a mortality rate in men three times that in women has been found. Furthermore, a decrease in testosterone has also been associated with aging and comorbidities such as obesity, diabetes mellitus, and cardiovascular diseases. In addition, testosterone has shown an inverse relationship with proinflammatory cytokines and neutrophil count and a direct relationship with lymphocyte count, demonstrating its immunomodulatory and protective effect [160].
ARDS is the main complication in patients with COVID-19 and is related to several of the biomarkers of disease severity, but it has also been reported that the pulmonary microbiota is significantly different between patients with and without ARDS. This microbiota modulates the immune system. It has been shown that the enrichment of the pulmonary microbiota with intestinal bacteria was related to elevated inflammatory biomarkers in plasma. In general, patients with severe disease showed the presence of high concentrations of Bacteroides, Enterobacteriaceae, and Lachnospiraceae associated with the intestine [161].
Monitoring signatures of biomarkers with biosensors in the context of precision medicine
Precision medicine, also known as personalized medicine, seeks to tailor disease prevention and treatment by considering genetic differences in individuals. Age-, sex-, and race-associated factors have been vital in predicting susceptibility to disease [162]. Each individual has variations in biomarkers at different molecular levels that may indicate a predisposition to disease or increased disease severity. These biomarkers range from DNA and RNA to functional proteins and metabolic molecules characteristic of multiple molecular events specific to each disease, and more than a single biomarker is required to support a more accurate prediction and prognosis. In this context, multiparametric detection at different molecular levels offers the best alternative for developing personalized medicine [163].
COVID-19 is an illustrative example of the application of precision medicine for prevention and treatment. Different biomarkers like those mentioned in the previous section are expressed differently in each individual. For example, the ACE2 receptor, found in the respiratory system and other places in the human body such as the eye and epithelial and gastrointestinal cells, is expressed in greater density in bronchial cells in men and is highest in adulthood but begins to decline with aging [164]. In addition, elevated levels of this receptor have been observed in lung cells of smokers and people with chronic obstructive pulmonary disease, and low levels in people with diabetes mellitus and heart disease [25, 164].
The ACE2 protein can vary in each population, and each variant has a different degree of binding affinity for protein S and the level of ACE2 expression in cells [116]. It has been shown that high levels of circulating ACE2 decrease the risk of infection because this soluble protein acts as a decoy for SARS-CoV-2 [116]. In addition, although high levels of ACE2 correlate with susceptibility to infection, low levels of this protein may increase the virus pathogenicity, as it increases the concentrations of angiotensin II in the body, which may also increase the inflammatory reaction and hypercoagulation processes [116, 164]. This makes the ACE2 protein an excellent biomarker for assessing disease prevention, severity, and treatment.
On the other hand, the gene cluster on chromosome 3p21.31 was identified as the main genetic susceptibility locus in patients with respiratory failure, as it encodes several chemokine receptors that directly affect the body's immune response [126]. Its rs11385942 variant in particular was found in a higher proportion of patients who required mechanical ventilation. Furthermore, the effect of SNP rs657152 A or C at locus 9q34.2, related to the ABO blood group, showed an increased risk of infection in people with blood group A and a protective effect in those with blood group O [165]. Also, the relationship between severe disease and pre-existing comorbidities such as obesity, diabetes, and cardiovascular disease has been documented. For example, type 2 diabetes and obesity are metabolic disorders that present immune dysfunction, accumulating a more significant number of immune cells and increasing the state of inflammation, which directly affects disease susceptibility and severity. In addition, high cholesterol levels indirectly facilitate viral entry, and low HDL levels increase disease severity [166].
Vitamin D deficiency was also a determining factor in susceptibility to infection. Vitamin D can enter the body from sunlight, food, or supplements. While vitamin D3 is obtained when ultraviolet B rays convert the precursor 7-dehydrocholesterol present in the skin, vitamin D2 can be obtained from milk, cereals, fish, or supplements. They are transformed in the liver into 25-hydroxyvitamin D—25(OH)D—present in serum and used as an indicator of vitamin D deficiency [167]. A strong association has been reported between low levels of this vitamin and SARS-CoV-2 infection [168, 169]. Infection-susceptible populations with older people from Italy, Switzerland, and Spain, or ethnic groups with darker skin, such as African Americans, showed lower serum 25(OH)D concentrations [167]. An effect of vitamin D on the severity of the disease has also been demonstrated, since this vitamin increases innate immunity, decreasing the generation of proinflammatory cytokines and consequently avoiding the cytokine storm [167]. Vitamin D deficiency was also associated with a higher prevalence of vascular disease and hypertension and elevated levels of FT and cTn. Normal levels were above 20 ng/mL, with an increased risk of hospitalization with concentrations below this level [168].
Finally, many polymorphisms have been reported in genes related to disease severity. These may modulate the susceptibility of individuals and the severity of the disease [170]. Table 2 shows several reported variants.
Conventionally ELISA-based immunoassays have been used to detect different biomarkers, and there are many commercially available ELISA kits (see Fig. 2c). However, developing biosensors for these biomarkers offers advantages over conventional methods in covering clinically prescribed ranges and offering portability and multiplexing possibilities of great utility for clinical use. Although ELISA is a gold standard technique, commercially available ELISA kits typically have sensitivity down to picograms per milliliter. In contrast, biosensors may have a wider linear working range, higher sensitivity, and lower limits of detection (LOD) [174]. These autonomous devices integrate materials and biomolecules coupled with transducers that transform the physical, chemical, or biological interaction of the bioreceptors and the analyte into a quantifiable signal for the selective and specific detection of molecular targets (analytes) [175,176,177,178,179]. These devices are characterized by high sensitivity, low LOD, and high specificity derived from the combination of the properties of nanomaterials and biological recognition systems [180]. In addition, they can be miniaturized and integrated into portable, multiparametric analysis systems using very small reagent and sample volumes, making assays fast, simple, and easy to implement even for nonexpert personnel [177].
Biosensors consist of a transducer, a biological component called a recognition element, and a portable reading device [176, 181, 182], as shown in Fig. 3. The biological components are immobilized on the surface of the transducers and interact with high affinity with the target molecule. The sensor generates a physicochemical signal that is converted into a measurable and quantifiable signal [176], which is sent to a processing system for amplification and analysis [177, 183]. Biological receptors are recognition elements that can be enzymes, proteins, antibodies, nucleic acids, cells, tissues, or receptor molecules [184]. These molecules are responsible for giving specificity to the biosensor and must be in direct contact with the transducer [179, 185]. Depending on the recognition element, biosensors can be catalytic, including enzymes, microorganisms, organelles, cells, or tissues and based on affinity, including antibodies, nucleic acids, proteins, peptides, and aptamers [176, 177]. Transducers offer different sensing and signal conversion strategies, classified as electrochemical, optical, mass, thermoelectric, piezoelectric, and calorimetric [177].
Components of a biosensor. The first component is the detection system that includes the transducer that can be modified with different nanomaterials and biological receptors to obtain high sensitivity and specificity. In some cases, it can also have amplification systems. The second component is the amplification and processing system for data visualization and analysis. This equipment can be portable and can have the capacity to analyze multiple samples at the same time. The sample can come from cell cultures, patients, food, or an environmental matrix and contains the analyte of interest. Modified from reference [177]
Electrochemical biosensors are widely explored due to their amenability to miniaturization, offering portability and thus new opportunities for POC detection. These biosensors measure physicochemical or biological bioreceptor–target interactions by changes in electrical properties at the electrode–solution interface [177, 178]. Advances in their development include modifying carbon, gold, and platinum-based electrodes with a wide variety of nanomaterials to improve their electroanalytical properties [186], increasing the surface area-to-volume ratio, creating stable and favorable microenvironments for the maintenance of the analytical biomolecule's structure, reducing LOD and response times and increasing the biosensor stability. In addition, these biosensors can be multiplexed to detect several biomarkers simultaneously, which brings us closer to their application in personalized medicine (see Fig. 2d).
Detection of SARS-CoV-2 based on electrochemical biosensors
Over the past 2 years, various strategies have been investigated for the detection of SARS-CoV-2, preferably at the POC in a rapid and ultrasensitive manner. Among the electrochemical biosensors, Seo et. al. [187] reported a field-effect transistor (FET)-based biosensor device to detect the S protein of the SARS-CoV-2, which was developed within months of the onset of the pandemic. After applying an input voltage, FET-based biosensors translated the biological signal into an electrical signal generated in a channel. The channel was coated with graphene sheets due to its high electrical conductivity, high carrier mobility, large surface area, and a capture antibody interacting specifically with S protein immobilized on the graphene sheets. The output signal was measured from FET transfer (current–voltage [I–V] curves), showing a LOD of 1 fg/mL in phosphate-buffered saline and 2.42 x 102 copies/mL in culture medium and patient samples, with the great advantage of being adaptable for the diagnosis of other emerging diseases.
Vadlamani et. al. [188] developed an electrochemical biosensor based on TiO2 nanotubes functionalized with cobalt (Co-TNTs), making it inexpensive, simple, cost-effective, and highly sensitive. The detection was based on forming a Co-S protein complex at a specific bias voltage due to Co ion reduction and S protein oxidation. This biosensor was designed to detect the RBD from protein S by amperometry with LOD of 12 nM, within a broad linear detection region and in only 30 s.
Fabiani et. al. [189] developed an electrochemical immunoassay to detect S and N proteins of the virus in saliva, using magnetic beads modified with an anti-mouse IgG as a carrier to immobilize a monoclonal capture antibody. A polyclonal antibody coupled to a secondary antibody conjugated with alkaline phosphatase was used to produce the signal. Detection was on screen-printed electrodes modified with carbon black nanomaterial by differential-pulse voltammetry (DPV) using a portable potentiostat. The biosensor detected S and N proteins of the virus in a buffer solution and saliva with LOD of 19 ng/mL and 8 ng/mL, respectively, in only 30 min, so the authors considered it to hold potential for commercialization. However, studies are required to improve the signal-to-noise ratio of the device.
Zhao et. al. [190] designed an electrochemical biosensor based on calixarene-functionalized graphene oxide and a Au@Fe3O4 nanocomposite to detect SARS-CoV-2 genetic material using a supersandwich strategy (capture probe coupled to Au@ Fe3O4 and the signal probe coupled to the functionalized graphene oxide) by DPV. This biosensor demonstrated high specificity and selectivity in in silico tests and real samples, with 200 copies/mL LOD. This biosensor was also integrated with a smartphone for POC analysis of the results.
Idili et. al [191] developed an aptamer-based electrochemical sensor for the rapid, sensitive, and reagent-free detection of the SARS-CoV-2 S protein. The sensor response was produced by a conformational change induced by the binding of the modified aptamer to a methylene blue derivative immobilized on a gold electrode surface. This response was so fast that it was able to recognize the target in a single step within 15 s, over a range of S protein concentrations from 760 pg/mL to 76 ng/mL.
An electrochemical immunosensor for detecting SARS-CoV-2 was recently developed by our group [22]. The sandwich-type immunosensor was based on magnetic particles that take advantage of the high-affinity interaction of the spike protein with the ACE2 protein and use a poly-horseradish peroxidase (HRP) enzyme complex as an amplification system. The reaction was followed by chronoamperometry after confining the particles at screen-printed gold electrodes, achieving a LOD of 22.5 ng/mL with only 5 μL of samples in a pocket potentiostat (see Fig. 4a). Our group [21] also developed the first electrochemical biosensor based on peptides immobilized on screen-printed gold electrodes for the straightforward and specific detection of untagged SARS-CoV-2 protein S by electrochemical impedance spectroscopy (EIS). The device demonstrated a LOD of 18.2 ng/mL protein S and 0.01 copies/mL of lysed particles (see Fig. 4b), concentrations of clinical relevance, and in only 15 min. Remarkably, both devices detected the SARS-CoV-2 in samples positive for the virus by RT-PCR and did not show a measurable signal in samples from healthy individuals. This highlights the potential of the as-developed biosensors to detect protein S from SARS-CoV-2 and viral particles from clinical samples. And finally, our group developed an electrochemical genosensor based on magnetic particles modified with thiolated capture probes (see Fig. 4c), which detects viral RNA sandwiched with biotinylated signal probes modified with enzyme complexes, achieving a LOD of 807 fM, and high specificity to discriminate SARS-CoV, MERS, and HKU1 homologous viruses [23]. These examples demonstrate the great advantages of applying electrochemical biosensors to biomarkers of different molecular levels in SARS-CoV-2 infection as a diagnostic component in personalized COVID-19 medicine.
Multilevel detection of SARS-CoV-2 with (a) electrochemical immunosensor based on magnetic beads and the spike-ACE2 complex [22], (b) first peptide-based impedimetric biosensor [21] for protein S detection, and (c) electrochemical genosensor based on magnetic beads for RNA detection [23]. Reproduced with permission. Copyright © 2022, Elsevier B.V.
Although significant advances have been made in biosensors, relatively few examples have been reported for SARS-CoV-2; some are summarized in Table 3.
Biosensors for prognosis and prediction of the course of COVID-19
As described above, the biomarkers most frequently related to susceptibility to infection were vitamin D and ACE2 protein, while for disease severity they were lymphocyte count (lymphopenia), PCT, IL6, and CrP, and for increased risk of death, the D-dimer, cTn, and LDH biomarkers [154]. However, electrochemical biosensors that detect various biomarkers at different molecular levels for diagnosis and prediction of the course of COVID-19 are scarce, due to the novelty of the SARS-CoV-2. Nevertheless, it is important to highlight the work of Prof. Gao's laboratory [48], which developed a low-cost, portable, and wireless multiplexed biosensor platform that enabled the rapid and ultrasensitive detection of three COVID-19-specific biomolecules. Furthermore, the device determined not only viral antigen nucleocapsid proteins (indicative of viral infection) and IgG and IgM antibodies (immune response) that provide information on the disease stage, but also CrP as an indicator of disease severity (see Fig. 5). Bioreceptors were immobilized on laser-etched graphene electrodes and measured by DPV and open-circuit potential–electrochemical impedance spectroscopy (OCP-EIS). Blood and saliva samples were analyzed, showing a highly selective and rapid response, between 1 and 10 min in relevant physiological ranges. This is an excellent example of the benefits of implementing electrochemical biosensors at different molecular levels in personalized medicine since, in addition to detecting SARS-CoV-2 infection, the platform indicates the progression and severity of the disease.
Graphene-based RapidPlex multiplexed electrochemical platform for detecting nucleocapsid protein, IgG and IgM immunoglobulins, and CrP [48]. Reproduced with permission. Copyright © 2022, Elsevier B.V.
Khayamian et. al. [229] developed an electrochemical biosensor for cytokine storm tracking in COVID-19 patients using graphene-modified copper electrodes and EIS. It must be clarified that they were not interested in specific biomarker detection (only 50%) but in indicating disease severity with biomarkers of inflammation in blood samples, so they did not use bioreceptors in the detection. And finally, Jagannath et al. [49] developed an electrochemical biosensor for the detection of inflammatory proteins interferon-inducible protein 10 (IP-10), TNF-related apoptosis-inducing ligand (TRAIL), and CrP in sweat as a strategy for monitoring infections such as COVID-19 in a noninvasive and portable way, achieving LOD of 1 pg/mL (IP-10 and TRAIL) and 0.2 ng/mL (CrP). The biosensor was based on a zinc oxide semiconductor electrode system modified with specific monoclonal antibodies, detecting the biomarkers by EIS. Overall, these three electrochemical biosensors are the only multiparametric biosensors reported to date for detecting different biomarkers in the framework of COVID-19. This highlights an open avenue for developing multiplexed biosensor-based systems to fulfill the diagnosis, prognosis, and course of the disease requirements toward a new paradigm of personalized medicine for its integral management. Finally, blood is the primary source of detection of these biomarkers, but other less invasive sources have also been investigated, such as the detection of inflammatory biomarkers in tear film [230,231,232], or other biomarkers in saliva [163, 233].
Electrochemical biosensors for detecting multiple proteins involved in other pathologies
Although, at the moment, there are very few electrochemical biosensors developed for the detection of severity and progression biomarkers specific to COVID-19, it is important to mention electrochemical biosensors that have been developed in recent years for the detection of these biomarkers involved in other pathologies. For instance, researchers have used a conventional glassy carbon electrode (GCE), modified with p-aminobenzoic acid, p-aminothiophenol, and gold nanoparticles, to anchor IL-6-specific thiolated aptamers as a biomarker for colorectal cancer detection [234]. EIS was used to evaluate the biosensor, responding linearly from 5 pg/mL to 100 ng/mL and with a LOD of 1.6 pg/mL. Another study integrated a flow cell with electrodes modified with specific antibodies to monitor secreted IL-6 and TNF-α [235]. Both biomarkers were detected amperometrically with high sensitivity. On the other hand, microelectrodes have been shown to be potential candidates for POC testing or insertion into blood vessels for continuous monitoring of IL-6. Microelectrodes have been modified with IL-6-specific antibodies for antigen detection over a linear range between 20 and 100 pg/mL in 2.5 min. POC IL-6 testing can rapidly predict bacterial infections rather than wait one to three days for diagnosis by conventional detection methods, highlighting the advantages of electrochemical biosensors.
Yang and coworkers developed an electrochemiluminescence sensor for PCT [236], with N-(aminobutyl)-N-(ethylisoluminol) (ABEI) bound to ferritin as donor molecule and gold nanoparticles as acceptors. This donor–acceptor system generated a response in a linear range from 100 fg/mL to 50 ng/mL, with a LOD of 41 fg/mL. Although the system was very sensitive, the biosensor fabrication was somewhat complex and used multiple reagents, which increased its cost. Therefore, molybdenum/gold trioxide was used on reduced graphene oxide nanocomposites (MoO3/Au@rGO) to simplify the system. The nanocomposite then had excellent electrocatalytic activity toward hydrogen peroxide (H2O2), amplifying the generated signals [237], achieving detection of PCT with high sensitivity and a LOD of 2 fg/mL, better than that achieved by electrochemiluminescence.
An impedimetric sensor based on an interdigitated gold electrode modified with zinc oxide (ZnO) thin films was reported for PCT detection [238], involving only one set of antibodies, with easy fabrication and reduced cost. However, the sensor did not provide better analytical performance compared with the work of Yang et al. and Liu et al. Electrodes modified with Prussian blue analog nanocubes functionalized with toluidine blue were used as highly sensitive PCT detection platforms with extremely low LOD, thanks to the large surface area of the nanocubes where the antibodies were anchored [239]. Abbas et al. grew cupric tungstate (CuWO4) nanospheres in situ on graphene oxide (GO) and used it as a photoelectrochemical sensor for PCT detection [240] to improve the detection parameters. The sensor had a LOD of 0.15 pg/mL, and its excellent performance was attributed to the synergistic effects of CuWO4 and GO nanospheres, which formed an effective photoactive heterojunction, an essential requirement in a photoelectrochemical sensing platform.
A new biosensor was built using a GCE modified with gold nanoparticles and delaminated sulfur-doped MXene for PCT detection [241], with an improved LOD (2 fg/mL). In addition, to improve the analytical performance of the sensors, iron sulfide (Fe3S4) loaded with Pd nanoparticles was used for PCT detection, with a methodology similar to that of the MoO3/Au@rGO-based biosensor mentioned above, achieving a LOD of only 130 fg/mL and a wider linear range [242]. The most recent work on PCT detection compared the performance of a microfluidic device integrating a gold and a screen-printed carbon electrode (SPCE) for PCT immunodetection [243] based on magnetic beads. The microfluidic system-based method had a lower LOD than the SPCE without system integration, but the latter provided a more extensive working range.
Electrochemical detection of FT using quantum dots (QDs) functionalized with biosurfactants was reported [244], with a response in a linear detection range of 10 to 1500 ng/mL, covering the clinical range, by DPV and cyclic voltammetry (CV), with LOD of 3.8 and 6.0 ng/mL, respectively. The sensor performance was also evaluated with human serum samples, with satisfactory results. The transducer was then functionalized with white graphene QDs (hexagonal boron nitride) to improve its performance. The improved linear range and LOD were 10–2000 ng/L and 1.3 ng/mL, respectively [245]. Recently, an electrochemical ferritin sensor was reported on a graphene-modified paper [246]. Antibodies were bound to the electrode surface by EDC/NHS. The biosensor response was linear from 1.0 to 1000 ng/mL, with LOD of 0.19 ng/mL.
A FET-based sensor was fabricated in the most recent report on ferritin detection. The FET was modified with graphene and 1-pyrenebutanoic acid and succinimidyl ester to bind anti-ferritin antibodies, improving the linear range and LODs of the state of the art, with a response time of only 1 to 10 s [247]. Garg et al. recently reported a microfluidic system for continuous electrochemical detection of ferritin. The system involved a screen-printed electrode modified with amine-functionalized graphene oxide. Anti-ferritin antibodies were immobilized on the electrode surface, which selectively detected ferritin dynamically, albeit with a linear range and LODs that did not exceed previous work [248]. Table 4 reports some electrochemical biosensors developed in recent years to detect biomarkers involved in other pathologies.
Concluding remarks, remaining challenges, and perspectives
The detection of biomarkers at different molecular levels offers tremendous opportunities for personalized medicine applied to emerging diseases such as COVID-19, but is currently a significant challenge. Recently, the detection of biomarkers associated with this viral infection has been proposed as an alternative to advance toward a more accurate diagnosis of patients with low viral load and new opportunities for prognosis and determination of the severity of the disease. It has been shown that the infection produced by SARS-CoV-2 triggers several inflammatory factors and biochemical and hematological biomarkers that, depending on the route of infection followed by the virus, produce a significant inflammatory response. Thus, several inflammatory markers, including CrP, IL-6, PCT and FT, have been closely related to this infection, mainly to the cytokine storm in severe disease cases. In addition, it has been related to miRNA, genes and their variants, and even cells of the immune system that can predict the path followed by the disease and may be the key to designing a targeted treatment for each patient in a personalized manner.
Detection of the mentioned biomarkers is the initial step to achieving precision medicine for COVID-19 management, where electrochemical biosensors play a pivotal role. Electrochemical biosensors enjoy exceptional properties for monitoring biomarkers in a sensitive and specific manner, being amenable for implementation at the POC. They also present the outstanding versatility to be multiplexed, not only enabling the diagnosis of the disease but also elucidating the prognosis of patients, responding quickly to the possibility of severe disease and/or death, and finally, following up the post-COVID-19 sequelae remaining in many people. Furthermore, notwithstanding the advances in sequencing the human genome, its variations and prevalence have been highly significant, so it is necessary to continue studying the effects of polymorphisms or variants in the susceptibility and progression of COVID-19, providing specific targets for detection and monitoring. Therefore, it is critical to unite research efforts in biology, medicine, clinical science, metrology, data processing, deep learning algorithms, and artificial intelligence to advance the development of integrated tools for the prevention, diagnosis, monitoring, and treatment of the disease.
Despite the progress discussed above, there are still several challenges to address, such as the need for regulatory agencies to compare and test the validity of different detection platforms to maintain reliability and demonstrate their efficacy relative to the current standard of care. For example, electrochemical biosensors for SARS-CoV-2 with validated response to the latest circulating variants are still needed. Another critical challenge is demonstrating the clinical efficacy of these devices and the high benefit/cost ratio, as prevention and early diagnosis can significantly reduce costs associated with treating serious diseases. The final challenge would be to bring these outstanding achievements and results to the industry, moving us closer to the future of health and personalized medicine. Finally, further advances are also needed in developing multiplexed devices and monitoring several biomarkers simultaneously, allowing health personnel to make more rapid and precise decisions in treating patients. Furthermore, achieving a personalized treatment could improve response times, reducing side effects, drug resistance, and even the psychological effects that can impede recovery due to nonspecific treatment.
Overall, personalized medicine for COVID-19 and other diseases that currently claim millions of lives per year is the way to the future—taking advantage of electrochemical biosensors as simple tracking tools and versatile monitoring strategies would make it possible to implement unique and individualized approaches for prevention, diagnosis, and treatment of diseases, toward establishing a new paradigm of personalized medicine.
References
Weil AR. Precision Medicine. 2018;37(5):687. https://doi.org/10.1377/hlthaff20180520.
König IR, Fuchs O, Hansen G, Von Mutius E, Kopp MV. What is precision medicine? Review precision medicine. Eur Respir J [Internet]. 2017;50:1700391. https://doi.org/10.1183/13993003.00391-2017.
Alonso SG, de la Torre Díez I, Zapiraín BG. Predictive, Personalized, Preventive and Participatory (4P) Medicine Applied to Telemedicine and eHealth in the Literature. J Med Syst 2019 435 [Internet]. 2019;43(5):1–10. Available from: https://link.springer.com/article/10.1007/s10916-019-1279-4.
Battle A, Mostafavi S, Zhu X, Potash JB, Weissman MM, McCormick C, et al. Characterizing the genetic basis of transcriptome diversity through RNA-sequencing of 922 individuals. Genome Res [Internet]. 2014;24(1):14–24 Available from: https://genome.cshlp.org/content/24/1/14.full.
Cenik C, Cenik ES, Byeon GW, Grubert F, Candille SI, Spacek D, et al. Integrative analysis of RNA, translation, and protein levels reveals distinct regulatory variation across humans. Genome Res [Internet]. 2015;25(11):1610–21 Available from: https://genome.cshlp.org/content/25/11/1610.full.
Wu L, Candille SI, Choi Y, Xie D, Jiang L, Li-Pook-Than J, et al. Variation and genetic control of protein abundance in humans. Nat 2013 4997456 [Internet]. 2013;499(7456):79–82. Available from: https://www.nature.com/articles/nature12223.
Priyadharshini VS, Teran LM. Personalized Medicine in Respiratory Disease: Role of Proteomics. Adv Protein Chem Struct Biol. 2016;1(102):115–46.
Xie J, Lee S, Chen X. Nanoparticle-based theranostic agents. Adv Drug Deliv Rev. 2010;62(11):1064–79.
Cruz JA, Wishart DS. Applications of Machine Learning in Cancer Prediction and Prognosis: https://doi.org/10.1177/117693510600200030 [Internet]. 2017 Oct 4;2:59–77. Available from: https://journals.sagepub.com/doi/full/10.1177/117693510600200030.
Williams MS. Early Lessons from the Implementation of Genomic Medicine Programs. https://doi.org/10.1146/annurev-genom-083118-014924 [Internet]. 2019 Aug 30;20:389–411. Available from: https://www.annualreviews.org/doi/abs/10.1146/annurev-genom-083118-014924.
Altmann A, Beerenwinkel N, Sing T, Savenkov I, Däumer M, Kaiser R, et al. Improved Prediction of Response to Antiretroviral Combination Therapy using the Genetic Barrier to Drug Resistance: https://doi.org/10.1177/135965350701200202 [Internet]. 2006 Feb 1; Available from: https://journals.sagepub.com/doi/abs/10.1177/135965350701200202.
Kelkar SS, Reineke TM. Theranostics: Combining imaging and therapy. Bioconjug Chem [Internet]. 2011;22(10):1879–903 Available from: https://pubs.acs.org/doi/abs/10.1021/bc200151q.
Kamps R, Brandão RD, van den Bosch BJ, Paulussen ADC, Xanthoulea S, Blok MJ, et al. Next-Generation Sequencing in Oncology: Genetic Diagnosis, Risk Prediction and Cancer Classification. Int J Mol Sci 2017, Vol 18, Page 308 [Internet]. 2017 Jan 31;18(2):308. Available from: https://www.mdpi.com/1422-0067/18/2/308/htm
Yahata N, Kasai K, Kawato M. Computational neuroscience approach to biomarkers and treatments for mental disorders. Psychiatry Clin Neurosci [Internet]. 2017;71(4):215–37.
Jørgensen JT. The current landscape of the FDA approved companion diagnostics. Transl Oncol. 2021;14(6):101063.
Mancinelli L, Cronin M, Sadée W. Pharmacogenomics: The promise of personalized medicine. AAPS PharmSci 2002 21 [Internet]. 2000;2(1):29–41. Available from: https://link.springer.com/article/10.1208/ps020104
Fernández M, Orozco J. Advances in Functionalized Photosensitive Polymeric Nanocarriers. Polym. 13(15):2464 Available from: https://www.mdpi.com/2073-4360/13/15/2464/htm.
Mena-Giraldo P, Pérez-Buitrago S, Londoño-Berrío M, Ortiz-Trujillo IC, Hoyos-Palacio LM, Orozco J. Photosensitive nanocarriers for specific delivery of cargo into cells. Sci Reports 2020 101 [Internet]. 10(1):1–12 Available from: https://www.nature.com/articles/s41598-020-58865-z.
Harrison AG, Lin T, Wang P. Mechanisms of SARS-CoV-2 Transmission and Pathogenesis. Trends Immunol. 2020;41(12):1100–15.
Jackson CB, Farzan M, Chen B, Choe H. Mechanisms of SARS-CoV-2 entry into cells. Nat Rev Mol Cell Biol. 2021;23(1):3–20 Available from: https://www.nature.com/articles/s41580-021-00418-x.
Soto D, Orozco J. Peptide-based simple detection of SARS-CoV-2 with electrochemical readout. Anal Chim Acta [Internet]. 2022;1(1205):339739 Available from: https://linkinghub.elsevier.com/retrieve/pii/S0003267022003105.
Vásquez V, Navas M-C, Jaimes JA, Orozco J. SARS-CoV-2 electrochemical immunosensor based on the spike-ACE2 complex. Anal Chim Acta [Internet]. 2022;339718. Available from: https://linkinghub.elsevier.com/retrieve/pii/S0003267022002896
Cajigas S, Alzate D, Fernández M, Muskus C, Orozco J. Electrochemical genosensor for the specific detection of SARS-CoV-2. Talanta [Internet]. 2022;16:123482 Available from: https://linkinghub.elsevier.com/retrieve/pii/S0039914022002788.
Wan J, Xing S, Ding L, Wang Y, Gu C, Wu Y, et al. Human-IgG-Neutralizing Monoclonal Antibodies Block the SARS-CoV-2 Infection. Cell Rep [Internet]. 2020;32(3):107918 Available from: https://www.sciencedirect.com/science/article/pii/S2211124720308998#:~:text=Summary,-andlight-chainfragments.
Zhang L, Guo H. Biomarkers of COVID-19 and technologies to combat SARS-CoV-2. Adv Biomark Sci Technol [Internet]. 2020;1(2):1–23 Available from: https://www.sciencedirect.com/science/article/pii/S2543106420300016.
Collivignarelli MC, Collivignarelli C, Carnevale Miino M, Abbà A, Pedrazzani R, Bertanza G. SARS-CoV-2 in sewer systems and connected facilities. Process Saf Environ Prot. 2020;143:196–203.
Shereen MA, Khan S, Kazmi A, Bashir N, Siddique R. COVID-19 infection: Origin, transmission, and characteristics of human coronaviruses. J Adv Res [Internet]. 2020;24:91–8. https://doi.org/10.1016/j.jare.2020.03.005.
Wu A, Peng Y, Huang B, Ding X, Wang X, Niu P, et al. Genome Composition and Divergence of the Novel Coronavirus (2019-nCoV) Originating in China. Cell Host Microbe [Internet]. 2020;27(3):325–8. https://doi.org/10.1016/j.chom.2020.02.001.
Luo Z, Ang MJY, Chan SY, Yi Z, Goh YY, Yan S, et al. Combating the Coronavirus Pandemic: Early Detection, Medical Treatment, and a Concerted Effort by the Global Community. Theatr Res Int. 2020;16(2020):1–35 Available from: https://spj.sciencemag.org/journals/research/2020/6925296/.
Wrobel AG, Benton DJ, Xu P, Roustan C, Martin SR, Rosenthal PB, et al. SARS-CoV-2 and bat RaTG13 spike glycoprotein structures inform on virus evolution and furin-cleavage effects. Nat Struct Mol Biol [Internet]. 2020;27(8):763–7 Available from: https://www.nature.com/articles/s41594-020-0468-7.
Walls AC, Park YJ, Tortorici MA, Wall A, McGuire AT, Veesler D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell [Internet]. 2020;181(2):281-292.e6. Available from: https://doi.org/10.1016/j.cell.2020.02.058
Yang J, Petitjean S, Derclaye S, Koehler M, Zhang Q, Dumitru A, et al. Molecular interaction and inhibition of SARS-CoV-2 binding to the ACE2 receptor. Res Sq [Internet]. 2020; Available from: https://www.researchsquare.com/article/rs-30468/v1.
WHO Coronavirus (COVID-19) Dashboard | WHO Coronavirus (COVID-19) Dashboard With Vaccination Data [Internet]. Available from: https://covid19.who.int/.
Mathieu E, Ritchie H, Ortiz-Ospina E, Roser M, Hasell J, Appel C, et al. Coronavirus Pandemic (COVID-19). Our World Data [Internet]. 2020;5(7):947–53 Available from: https://ourworldindata.org/coronavirus.
Chams N, Chams S, Badran R, Shams A, Araji A, Raad M, et al. COVID-19: A Multidisciplinary Review. Front Public Health. 2020;8(July):1–20.
Rabaan AA, Al-Ahmed SH, Haque S, Sah R, Tiwari R, Singh Malik Y, et al. SARS-CoV-2, SARS-CoV, and MERS-CoV: a comparative overview [Internet]. 2020. Available from: https://www.infezmed.it/media/journal/Vol_28_2_2020_7.pdf
Cubuk J, Alston JJ, Incicco JJ, Singh S, Stuchell-Brereton MD, Ward MD, et al. The SARS-CoV-2 nucleocapsid protein is dynamic, disordered, and phase separates with RNA. Nat Commun 2021 121 [Internet]. 2021 Mar 29;12(1):1–17. Available from: https://www.nature.com/articles/s41467-021-21953-3.
Kubina R, Dziedzic A. Molecular and serological tests for COVID-19. A comparative review of SARS-CoV-2 coronavirus laboratory and point-of-care diagnostics. Diagnostics. 2020;10(6).
Malik YA. Properties of Coronavirus and SARS-CoV-2. 2020 [cited 2020 Nov 5];3–11. Available from: http://www.mjpath.org.my/2020/v42n1/properties-of-coronavirus.pdf.
Sino Biological. Human Coronavirus Spike [Internet]. 2020. Available from: https://www.sinobiological.com/research/virus/human-coronavirus-spike.
Esakandari H, Nabi-Afjadi M, Fakkari-Afjadi J, Farahmandian N, Miresmaeili SM, Bahreini E. A comprehensive review of COVID-19 characteristics. Biol Proced Online. 2020;22(1):1–10.
Wang S, Qiu Z, Hou Y, Deng X, Xu W, Zheng T, et al. AXL is a candidate receptor for SARS-CoV-2 that promotes infection of pulmonary and bronchial epithelial cells. Cell Res 2021 312 [Internet]. 2021 Jan 8;31(2):126–40. Available from: https://www.nature.com/articles/s41422-020-00460-y.
Delgado JM, Duro N, Rogers DM, Tkatchenko A, Pandit SA, Varma S. Molecular basis for higher affinity of SARS-CoV-2 spike RBD for human ACE2 receptor. Proteins Struct Funct Bioinforma [Internet]. 2021;89(9):1134–44 Available from: https://onlinelibrary.wiley.com/doi/full/10.1002/prot.26086.
Lu J, Sun PD. High affinity binding of SARS-CoV-2 spike protein enhances ACE2 carboxypeptidase activity. J Biol Chem [Internet]. 2020 Dec 25;295(52):18579–88. Available from: http://www.jbc.org/article/S0021925817507206/fulltext.
A. Al-Azzawi M, A. Sakr M. Co-Evolution between New Coronavirus (SARS-CoV-2) and Genetic Diversity: Insights on Population Susceptibility and Potential Therapeutic Innovations. In: Genetic Diversity [Working Title] [Internet]. IntechOpen; 2020. Available from: www.intechopen.com.
Wang Q, Zhang Y, Wu L, Niu S, Song C, Zhang Z, et al. Structural and Functional Basis of SARS-CoV-2 Entry by Using Human ACE2. Cell. 2020;181(4):894–904.e9.
Yan R, Zhang Y, Li Y, Xia L, Guo Y, Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science (80- ) [Internet]. 2020 Mar 27;367(6485):1444–8. Available from: /pmc/articles/PMC7164635/?report=abstract
Torrente-Rodríguez RM, Lukas H, Tu J, Xu C, Rossiter HB, Gao W. SARS-CoV-2 RapidPlex: A Graphene-Based Multiplexed Telemedicine Platform for Rapid and Low-Cost COVID-19 Diagnosis and Monitoring. Matter [Internet]. 2020. https://doi.org/10.1016/j.matt.2020.09.027.
Jagannath B, Pali M, Lin KC, Sankhala D, Naraghi P, Muthukumar S, et al. Novel Approach to Track the Lifecycle of Inflammation from Chemokine Expression to Inflammatory Proteins in Sweat Using Electrochemical Biosensor. Adv Mater Technol [Internet]. 2022;2101356. Available from: https://onlinelibrary.wiley.com/doi/full/10.1002/admt.202101356.
Koteswara RV. Point of Care Diagnostic Devices for Rapid Detection of Novel Coronavirus (SARS-nCoV19) Pandemic: A Review. Front Nanotechnol. 2021;2:22.
Jalandra R, Yadav AK, Verma D, Dalal N, Sharma M, Singh R, et al. Strategies and perspectives to develop SARS-CoV-2 detection methods and diagnostics. Biomed Pharmacother. 2020;129:110446.
Green K, Graziadio S, Turner P, Fanshawe T, Allen J. Molecular and antibody point-of-care tests to support the screening, diagnosis and monitoring of COVID-19; Available from: www.cebm.net/oxford-covid-19/.
Corman VM, Haage VC, Bleicker T, Schmidt ML, Mühlemann B, Zuchowski M, et al. Comparison of seven commercial SARS-CoV-2 rapid point-of-care antigen tests: a single-centre laboratory evaluation study. The Lancet Microbe. 2021;2(7):e311–9.
Liu G, Jiang C, Lin X, Yang Y. Point-of-care detection of cytokines in cytokine storm management and beyond: Significance and challenges. View [Internet]. 2021;2(4):20210003 Available from: https://onlinelibrary.wiley.com/doi/full/10.1002/VIW.20210003.
Stenken JA, Poschenrieder AJ. Bioanalytical chemistry of cytokines – A review. Anal Chim Acta. 2015;853(1):95–115.
Bains S, Anyaeche C, Wyatt A, Coker O, Bolodeoku J. Evaluation of Point of Care Test (POCT), i-CHROMA Serum C-Reactive Protein (CRP) Assay and Microalbumin Urine (MAU) Methods. Ann Clin Lab Res [Internet]. 2017;5(3):0–0. Available from: https://www.itmedicalteam.pl/articles/evaluation-of-point-of-care-test-poct-ichroma-serum-creactive-protein-crp-assay-and-microalbumin-urine-mau-methods-102171.html.
ichromaTM II - Boditech Med [Internet]. Available from: https://www.boditech.co.kr/en/product/instruments/id/4.
Chandra P, Suman P, Airon H, Mukherjee M, Kumar P. Prospects and advancements in C-reactive protein detection. World J Methodol [Internet]. 2014;4(1):1 Available from: /pmc/articles/PMC4145575/.
Pohanka M. Diagnoses Based on C-Reactive Protein Point-of-Care Tests. Biosens 2022, Vol 12, Page 344 [Internet]. 12(5):344. Available from: https://www.mdpi.com/2079-6374/12/5/344/htm.
Samsudin I, Vasikaran SD. Clinical Utility and Measurement of Procalcitonin. Clin Biochem Rev [Internet]. 2017;38(2):59 Available from: /pmc/articles/PMC5759088/.
Li C, Huang Y, Xu Y. Determining procalcitonin at point-of-care; A method comparison study of four commercial PCT assays. Pract Lab Med. 2021;25:e00214.
Riedel S, Melendez JH, An AT, Rosenbaum JE, Zenilman JM. Procalcitonin as a Marker for the Detection of Bacteremia and Sepsis in the Emergency Department. Am J Clin Pathol [Internet]. 2011;135(2):182–9 Available from: https://academic.oup.com/ajcp/article/135/2/182/1760294.
Garcia-Casal MN, Peña-Rosas JP, Urrechaga E, Escanero JF, Huo J, Martinez RX, et al. Performance and comparability of laboratory methods for measuring ferritin concentrations in human serum or plasma: A systematic review and meta-analysis. PLoS One [Internet]. 2018;13(5). Available from: /pmc/articles/PMC5933730/.
Garcia-Casal MN, Pasricha SR, Martinez RX, Lopez-Perez L, Peña-Rosas JP. Serum or plasma ferritin concentration as an index of iron deficiency and overload. Cochrane Database Syst Rev [Internet]. 2021;2021(5). Available from: /pmc/articles/PMC8142307/.
Ding S, Ma J, Song X, Dong X, Xie L, Song X, et al. Diagnostic Accuracy of Procalcitonin, Neutrophil-to-Lymphocyte Ratio, and C-Reactive Protein in Detection of Bacterial Infections and Prediction of Outcome in Nonneutropenic Febrile Patients with Lung Malignancy. J Oncol. 2020;2020.
Bui HN, Bogers JPAM, Cohen D, Njo T, Herruer MH. Evaluation of the performance of a point-of-care method for total and differential white blood cell count in clozapine users. Int J Lab Hematol. 2016;38(6):703–9.
Luo J, Chen C, Li Q. White blood cell counting at point-of-care testing: A review. Electrophoresis [Internet]. 2020;41(16–17):1450–68 Available from: https://onlinelibrary.wiley.com/doi/full/10.1002/elps.202000029.
Kocyigit I, Eroglu E, Safa Kaynar A, Kocer D, Kargi S, Zararsiz G, et al. The association of endothelin-1 levels with renal survival in polycystic kidney disease patients. J Nephrol [Internet]. 2019;32:83–91. https://doi.org/10.1007/s40620-018-0514-2.
Rossi GP, Seccia TM, Albertin G, Pessina AC. Measurement of endothelin: clinical and research use.
Akl MR, Nagpal P, Ayoub NM, Prabhu SA, Gliksman M, Tai B, et al. Molecular and clinical profiles of syndecan-1 in solid and hematological cancer for prognosis and precision medicine. Oncotarget [Internet]. 2015;6(30):28693–715 Available from: https://www.oncotarget.com/article/4981/text/.
Weitz JI, Fredenburgh JC, Eikelboom JW. A Test in Context: D-Dimer. J Am Coll Cardiol [Internet]. 2017;70(19):2411–20 Available from: https://www.jacc.org/doi/10.1016/j.jacc.2017.09.024.
Neale D, Tovey C, Vali A, Davies S, Myers K, Obiako M, et al. Evaluation of the Simplify d-dimer assay as a screening test for the diagnosis of deep vein thrombosis in an emergency department. Emerg Med J [Internet] 2004;21(6):663–6. Available from: https://emj.bmj.com/content/21/6/663.
Marquardt U, Apau D. Point-of-care D-dimer testing in emergency departments. Emerg Nurse. 2015;23(5):29–35.
Excellence NI for H and C. High-sensitivity troponin tests for the early rule out of NSTEMI Diagnostics guidance. 2020; Available from: www.nice.org.uk/guidance/dg40.
Luo J, Li S, Xu M, Guan M, Yang M, Ren J, et al. Real-time detection of cardiac troponin I and mechanism analysis of AlGaAs/GaAs high electron mobility transistor biosensor. AIP Adv [Internet]. 2020;10(11):115205 Available from: https://aip.scitation.org/doi/abs/10.1063/5.0027553.
PL_Calmark_COVID19-LDH_EN_v503; Available from: www.calmark.se.
Wu Y, Lu C, Pan N, Zhang M, An Y, Xu M, et al. Serum lactate dehydrogenase activities as systems biomarkers for 48 types of human diseases. Sci Reports 2021 111 [Internet]. 2021;11(1):1–8. Available from: https://www.nature.com/articles/s41598-021-92430-6.
Faruqi S, Wilmot R, Wright C, Morice AH. Serum LDH in chronic cough: a potential marker of airway inflammation. Clin Respir J [Internet]. 2012;6(2):81–7 Available from: https://onlinelibrary.wiley.com/doi/full/10.1111/j.1752-699X.2011.00250.x.
Halvorsen CP, Olson L, Araújo AC, Karlsson M, Nguyễn TT, Khu DTK, et al. A rapid smartphone-based lactate dehydrogenase test for neonatal diagnostics at the point of care. Sci Reports 2019 91 [Internet]. 2019;9(1):1–8. Available from: https://www.nature.com/articles/s41598-019-45606-0.
Moreira FTC, Dutra RAF, Noronha JP, Sales MGF. Novel sensory surface for creatine kinase electrochemical detection. Biosens Bioelectron. 2014;56:217–22.
Bruins Slot MHE, Van Der Heijden GJMG, Stelpstra SD, Hoes AW, Rutten FH. Point-of-care tests in suspected acute myocardial infarction: A systematic review. Int J Cardiol. 2013;168(6):5355–62.
Lindström M, Valkonen M, Tohmola N, Renkonen R, Strandin T, Vaheri A, et al. Plasma bradykinin concentrations during septic shock determined by a novel LC-MS/MS assay. Clin Chim Acta. 2019;493:20–4.
Zhou Y, Wang W, Wei R, Jiang G, Li F, Chen X, et al. Serum bradykinin levels as a diagnostic marker in cervical cancer with a potential mechanism to promote VEGF expression via BDKRB2. Int J Oncol [Internet]. 2019;55(1):131–41 Available from: http://www.spandidos-publications.com/10.3892/ijo.2019.4792/abstract.
Gangnus T, Burckhardt BB. Improving sensitivity for the targeted LC-MS/MS analysis of the peptide bradykinin using a design of experiments approach. Talanta. 2020;218:121134.
Dave VP, Ngo TA, Pernestig AK, Tilevik D, Kant K, Nguyen T, et al. MicroRNA amplification and detection technologies: opportunities and challenges for point of care diagnostics. Lab Investig 2018 994 [Internet]. 2018;99(4):452–69. Available from: https://www.nature.com/articles/s41374-018-0143-3
Ouyang T, Liu Z, Han Z, Ge Q. MicroRNA Detection Specificity: Recent Advances and Future Perspective. Anal Chem [Internet]. 2019;91(5):3179–86 Available from: https://pubs.acs.org/doi/full/10.1021/acs.analchem.8b05909.
Ye J, Xu M, Tian X, Cai S, Zeng S. Research advances in the detection of miRNA. J Pharm Anal. 2019;9(4):217–26.
Sun G, Xue J, Li L, Li X, Cui Y, Qiao B, et al. Quantitative determination of human serum testosterone via isotope dilution ultra-performance liquid chromatography tandem mass spectrometry. Mol Med Rep [Internet]. 2020;22(2):1576 Available from: /pmc/articles/PMC7339657/.
Ruan R, Deng X, Dong X, Wang Q, Lv X, Si C. Microbiota Emergencies in the Diagnosis of Lung Diseases: A Meta-Analysis. Front Cell Infect Microbiol. 2021;11:836.
Carney SM, Clemente JC, Cox MJ, Dickson RP, Huang YJ, Kitsios GD, et al. Methods in lung microbiome research. Am J Respir Cell Mol Biol [Internet]. 2020;62(3):283–99 Available from: www.atsjournals.org.
Dumont-Leblond N, Veillette M, Racine C, Joubert P, Duchaine C. Development of a robust protocol for the characterization of the pulmonary microbiota. Commun Biol 2021 41 [Internet]. 2021;4(1):1–9. Available from: https://www.nature.com/articles/s42003-021-01690-5
Xiao F, Burns KD. Measurement of Angiotensin Converting Enzyme 2 Activity in Biological Fluid (ACE2). Hypertension [Internet]. 2017;1527:101 Available from: /pmc/articles/PMC7121061/.
Rotondi M, Coperchini F, Ricci G, Denegri M, Croce L, Ngnitejeu ST, et al. Detection of SARS-COV-2 receptor ACE-2 mRNA in thyroid cells: a clue for COVID-19-related subacute thyroiditis. J Endocrinol Invest [Internet]. 2021;44(5):1085–90 Available from: https://link.springer.com/article/10.1007/s40618-020-01436-w.
Usami Y, Hirose K, Okumura M, Toyosawa S, Sakai T. Brief communication: Immunohistochemical detection of ACE2 in human salivary gland. Oral Sci Int [Internet]. 2021;18(2):101–4 Available from: https://onlinelibrary.wiley.com/doi/full/10.1002/osi2.1085.
DiaSys introduces Vitamin D test - DiaSys Diagnostic Systems GmbH [Internet]. Available from: https://www.diasys-diagnostics.com/company/news/details/news-article/diasys-introduces-vitamin-d-test/.
Albrecht K, Lotz J, Frommer L, Lackner KJ, Kahaly GJ. A rapid point-of-care assay accurately measures vitamin D. J Endocrinol Invest [Internet]. 2021;44(11):2485–92 Available from: https://link.springer.com/article/10.1007/s40618-021-01575-8.
Holick MF. Vitamin d status: measurement, interpretation and clinical application. Ann Epidemiol [Internet]. 2009;19(2):73 Available from: /pmc/articles/PMC2665033/.
Gallelli L, Michniewicz A, Cione E, Squillace A, Colosimo M, Pelaia C, et al. 25-Hydroxy Vitamin D Detection Using Different Analytic Methods in Patients with Migraine. J Clin Med [Internet]. 2019;8(6):895 Available from: /pmc/articles/PMC6617382/.
Lu Q, Nunez E, Lin C, Christensen K, Downs T, Carson DA, et al. A sensitive array-based assay for identifying multiple TMPRSS2:ERG fusion gene variants. Nucleic Acids Res [Internet]. 2008;36(20):e130. Available from: https://academic.oup.com/nar/article/36/20/e130/2902278.
Zhu F, Zhong Y, Ji H, Ge R, Guo L, Song H, et al. ACE2 and TMPRSS2 in human saliva can adsorb to the oral mucosal epithelium. J Anat [Internet]. 2022;240(2):398–409 Available from: https://onlinelibrary.wiley.com/doi/full/10.1111/joa.13560.
Priyadharshini VS, Teran LM. Role of respiratory proteomics in precision medicine. Precis Med Investig Pract Provid. 2020;1:255–61.
Kevadiya BD, Machhi J, Herskovitz J, Oleynikov MD, Blomberg WR, Bajwa N, et al. Diagnostics for SARS-CoV-2 infections. Nat Mater 2021 205 [Internet]. 2021 Feb 15;20(5):593–605. Available from: https://www.nature.com/articles/s41563-020-00906-z.
Mahapatra S, Chandra P. Clinically practiced and commercially viable nanobio engineered analytical methods for COVID-19 diagnosis. Biosens Bioelectron [Internet]. 2020;1(165):112361 Available from: https://www.sciencedirect.com/science/article/pii/S0956566320303559?dgcid=rss_sd_all.
Ravi N, Cortade DL, Ng E, Wang SX. Diagnostics for SARS-CoV-2 detection: A comprehensive review of the FDA-EUA COVID-19 testing landscape. Biosens Bioelectron. 2020;1(165):112454.
Sawano M, Takeshita K, Ohno H, Oka H. RT-PCR diagnosis of COVID-19 from exhaled breath condensate: a clinical study. J Breath Res [Internet]. 2021 Jul 1;15(3). Available from: https://pubmed.ncbi.nlm.nih.gov/34020435/
Chung YS, Lee NJ, Woo SH, Kim JM, Kim HM, Jo HJ, et al. Validation of real-time RT-PCR for detection of SARS-CoV-2 in the early stages of the COVID-19 outbreak in the Republic of Korea. Sci Reports 2021 111 [Internet]. 2021 Jul 20;11(1):1–8. Available from: https://www.nature.com/articles/s41598-021-94196-3.
PCR Test for COVID-19: What It Is, How Its Done, What The Results Mean [Internet].. Available from: https://my.clevelandclinic.org/health/diagnostics/21462-covid-19-and-pcr-testing.
Osterman A, Badell I, Basara E, Stern M, Kriesel F, Eletreby M, et al. Impaired detection of omicron by SARS-CoV-2 rapid antigen tests. Med Microbiol Immunol [Internet]. 2022;20:1–13 Available from: https://link.springer.com/article/10.1007/s00430-022-00730-z.
Routsias JG, Mavrouli M, Tsoplou P, Dioikitopoulou K, Tsakris A. Diagnostic performance of rapid antigen tests (RATs) for SARS-CoV-2 and their efficacy in monitoring the infectiousness of COVID-19 patients. Sci Reports 2021 111 [Internet]. 2021 Nov 24;11(1):1–9. Available from: https://www.nature.com/articles/s41598-021-02197-z.
Papadopoulou G, Manoloudi E, Repousi N, Skoura L, Hurst T, Karamitros T. Molecular and Clinical Prognostic Biomarkers of COVID-19 Severity and Persistence. Pathog 2022, Vol 11, Page 311 [Internet]. 2022 Mar 2;11(3):311. Available from: https://www.mdpi.com/2076-0817/11/3/311/htm.
Antibody (Serology) Testing for COVID-19: Information for Patients and Consumers | FDA [Internet].. Available from: https://www.fda.gov/medical-devices/coronavirus-covid-19-and-medical-devices/antibody-serology-testing-covid-19-information-patients-and-consumers.
Ejazi SA, Ghosh S, Ali N. Antibody detection assays for COVID-19 diagnosis: an early overview. Immunol Cell Biol [Internet]. 2021;99(1):21–33 Available from: https://pubmed.ncbi.nlm.nih.gov/32864735/.
Lou B, Li TD, Zheng SF, Su YY, Li ZY, Liu W, et al. Serology characteristics of SARS-CoV-2 infection since exposure and post symptom onset. Eur Respir J [Internet]. 2020 Aug 1;56(2). Available from: /pmc/articles/PMC7401320/.
Candel FJ, Barreiro P, Román JS, Abanades JC, Barba R, Barberán J, et al. Recommendations for use of antigenic tests in the diagnosis of acute SARS-CoV-2 infection in the second pandemic wave: attitude in different clinical settings. Rev Española Quimioter [Internet]. 2020;33(6):466 Available from: /pmc/articles/PMC7712344/.
Bortz RH, III, Florez C, Laudermilch E, Wirchnianski AS, Lasso G, et al. Single-Dilution COVID-19 Antibody Test with Qualitative and Quantitative Readouts. mSphere [Internet]. 2021 Apr 28;6(2). Available from: /pmc/articles/PMC8546701/.
Jahanafrooz Z, Chen Z, Bao J, Li H, Lipworth L, Guo X. An overview of human proteins and genes involved in SARS-CoV-2 infection. Gene. 2022;15(808):145963.
Alosaimi B, Hamed ME, Naeem A, Alsharef AA, AlQahtani SY, AlDosari KM, et al. MERS-CoV infection is associated with downregulation of genes encoding Th1 and Th2 cytokines/chemokines and elevated inflammatory innate immune response in the lower respiratory tract. Cytokine. 2020;1(126):154895.
Coperchini F, Chiovato L, Croce L, Magri F, Rotondi M. The cytokine storm in COVID-19: An overview of the involvement of the chemokine/chemokine-receptor system. Vol. 53, Cytokine and Growth Factor Reviews. Elsevier Ltd; 2020. p. 25–32.
Mahallawi WH, Khabour OF, Zhang Q, Makhdoum HM, Suliman BA. MERS-CoV infection in humans is associated with a pro-inflammatory Th1 and Th17 cytokine profile. Cytokine. 2018;104:8–13.
Ragab D, Salah Eldin H, Taeimah M, Khattab R, Salem R. The COVID-19 Cytokine Storm; What We Know So Far. Front Immunol [Internet]. 2020;11:1446 Available from: https://www.frontiersin.org/article/10.3389/fimmu.2020.01446/full.
Ye Q, Wang B, Mao J. The pathogenesis and treatment of the ‘Cytokine Storm” in COVID-19’ [Internet]. Vol. 80, Journal of Infection. W.B. Saunders Ltd; 2020. p. 607–13. https://doi.org/10.1016/j.jinf.2020.03.037.
Lingeswaran M, Goyal T, Ghosh R, Suri S, Mitra P, Misra S, et al. Inflammation, Immunity and Immunogenetics in COVID-19: A Narrative Review. Clin Biochem India [Internet]. 2020. https://doi.org/10.1007/s12291-020-00897-3.
Upadhyay J, Tiwari N, Ansari MN. Role of inflammatory markers in corona virus disease (COVID-19) patients: A review. Exp Biol Med [Internet]. 2020;245(15):1368–75 Available from: http://journals.sagepub.com/doi/10.1177/1535370220939477.
Kumar M, Taki K, Gahlot R, Sharma A, Dhangar K. A chronicle of SARS-CoV-2: Part-I - Epidemiology, diagnosis, prognosis, transmission and treatment. Sci Total Environ. 2020;734:139278.
Liu J, Li S, Liu J, Liang B, Wang X, Wang H, et al. Longitudinal characteristics of lymphocyte responses and cytokine profiles in the peripheral blood of SARS-CoV-2 infected patients. EBioMedicine. 2020;55:102763.
Mariappan V, Manoharan PS, Pajanivel R, Shanmugam L, Rao SR, Pillai AB. Potential biomarkers for the early prediction of SARS-COV-2 disease outcome. Microb Pathog. 2021;158:105057.
Peluso MJ, Lu S, Tang AF, Durstenfeld MS, Ho H, Goldberg SA, et al. Markers of immune activation and inflammation in individuals with post-acute sequelae of SARS-CoV-2 infection. medRxiv [Internet]. 2021 Jul 11;2021.07.09.21260287. Available from: https://www.medrxiv.org/content/10.1101/2021.07.09.21260287v1.
Tanaka T, Narazaki M, Kishimoto T, Il-6 in inflammation, Immunity, And disease. Cold Spring Harb Perspect Biol [Internet]. 2014;6(10):a016295 Available from: www.cshperspectives.org.
Matsuda T, Kishimoto T. Interleukin 6. In: Encyclopedia of Immunology [Internet]. Elsevier; 1998. p. 1458–61. Available from: https://linkinghub.elsevier.com/retrieve/pii/B0122267656003819.
Gabay C. Interleukin-6 and chronic inflammation. Arthritis Res Ther [Internet]. 2006;8(SUPPL. 2):S3 Available from: http://arthritis-research.biomedcentral.com/articles/10.1186/ar1917.
Khan A, Ali Z. Normal ranges for acute phase reactants (interleukin-6, tumour necrosis factor-alpha and C-reactive protein) in umbilical cord blood of healthy term neonates at the mount hope women’s hospital, Trinidad. West Indian Med J. 2014;63(5):465–9.
Grifoni E, Valoriani A, Cei F, Lamanna R, Gelli AMG, Ciambotti B, et al. Interleukin-6 as prognosticator in patients with COVID-19. J Infect [Internet]. 2020;81(3):452–82. https://doi.org/10.1016/j.jinf.2020.05.066.
Samprathi M, Jayashree M. Biomarkers in COVID-19: An Up-To-Date Review. Front Pediatr. 2021;8:972.
Seo HS. The role and clinical significance og high-sensitivity C-reactive protein in cardiovascular disease. Korean Circ J [Internet]. 2012; Available from: https://synapse.koreamed.org/upload/SynapseData/PDFData/0054kcj/kcj-42-151.pdf.
Nehring SM, Goyal A, Bansal P, Patel BC. C Reactive Protein. StatPearls [Internet]. 2021;65(5):237–44 Available from: https://www.ncbi.nlm.nih.gov/books/NBK441843/.
Vijayan AL, Ravindran S, Saikant R, Lakshmi S, Kartik R, Manoj G. Procalcitonin: A promising diagnostic marker for sepsis and antibiotic therapy. J Intensive Care [Internet] 2017;5(1):51. Available from: http://jintensivecare.biomedcentral.com/articles/10.1186/s40560-017-0246-8.
Lippi G, Plebani M. Procalcitonin in patients with severe coronavirus disease 2019 (COVID-19): A meta-analysis. Clin Chim Acta [Internet]. 2020;505:190–1 Available from: /pmc/articles/PMC7094472/.
Jin M, Khan AI. Procalcitonin: Uses in the Clinical Laboratory for the Diagnosis of Sepsis. Lab Med [Internet]. 2010;41(3):173–7 Available from: https://academic.oup.com/labmed/article-lookup/doi/10.1309/LMQ2GRR4QLFKHCH9.
Rubio-Sánchez R, Lepe-Balsalobre E. Viloria-Peñas M del M. Prognostic factors for the severity of SARS-CoV-2 infection. Adv Lab Med / Av en Med Lab [Internet]. 2021;2(2):253–8 Available from: https://www.degruyter.com/document/doi/10.1515/almed-2021-0017/html.
Zhou B, She J, Wang Y, Ma X. Utility of ferritin, procalcitonin, and C-reactive protein in severe patients with coronavirus disease 2019. 2020; Available from: https://assets.researchsquare.com/files/rs-23645/v1/4e6e3dbe-4004-4bb5-8c92-764d24bab8e4.pdf.
da Silva Antunes R, Mehta AK, Madge L, Tocker J, Croft M. TNFSF14 (LIGHT) exhibits inflammatory activities in lung fibroblasts complementary to IL-13 and TGF-β. Front Immunol. 2018;9(MAR):576.
Zhang L, Guo H. Biomarkers of COVID-19 and technologies to combat SARS-CoV-2. Adv Biomark Sci Technol [Internet]. 2020;2:1 Available from: /pmc/articles/PMC7435336/.
Perlin DS, Neil GA, Anderson C, Zafir-Lavie I, Roadcap L, Raines S, et al. CERC-002, a human anti-LIGHT mAb reduces respiratory failure and death in hospitalized COVID-19 ARDS patients. medRxiv [Internet]. 2021;2021.04.03.21254748. Available from: https://www.medrxiv.org/content/10.1101/2021.04.03.21254748v1.
Khalid A, Ali Jaffar M, Khan T, Abbas Lail R, Ali S, Aktas G, et al. Hematological and biochemical parameters as diagnostic and prognostic markers in SARS-COV-2 infected patients of Pakistan: a retrospective comparative analysis. https://doi.org/10.1080/1607845420211950898 [Internet]. 2021;26(1):529–42. Available from: https://www.tandfonline.com/doi/abs/10.1080/16078454.2021.1950898.
Maria Lagadinou, LastName EES, LastName NZ, LastName MM, Charalambos Gogos, LastName DV. Le Infezioni in Medicina. Le Infez Med [Internet]. 2020; Available from: https://www.infezmed.it/media/journal/Vol_28_suppl1_2020_14.pdf.
Ogawa F, Oi Y, Nakajima K, Matsumura R, Nakagawa T, Miyagawa T, et al. Temporal change in Syndecan-1 as a therapeutic target and a biomarker for the severity classification of COVID-19. Thromb J [Internet]. 2021;19(1):1–9 Available from: https://link.springer.com/articles/10.1186/s12959-021-00308-4.
Vidali S, Morosetti D, Cossu E, Luisa M, Luisi E, Pancani S, et al. D-dimer as an indicator of prognosis in SARS-CoV-2 infection: a systematic review. ERJ Open Res [Internet]. 2020;6(2):00260–2020 Available from: https://openres.ersjournals.com/content/6/2/00260-2020.
Yao Y, Cao J, Wang Q, Shi Q, Liu K, Luo Z, et al. D-dimer as a biomarker for disease severity and mortality in COVID-19 patients: A case control study. J Intensive Care [Internet]. 2020;8(1):1–11 Available from: https://jintensivecare.biomedcentral.com/articles/10.1186/s40560-020-00466-z.
Zhang L, Yan X, Fan Q, Liu H, Liu X, Liu Z, et al. D-dimer levels on admission to predict in-hospital mortality in patients with Covid-19. J Thromb Haemost [Internet]. 2020;18(6):1324–9 Available from: https://onlinelibrary.wiley.com/doi/full/10.1111/jth.14859.
Kavsak PA, Hammarsten O, Worster A, Smith SW, Apple FS. Cardiac Troponin Testing in Patients with COVID-19: A Strategy for Testing and Reporting Results. Clin Chem [Internet]. 2021;67(1):107–13 Available from: https://academic.oup.com/clinchem/article/67/1/107/5921202.
Lala A, Johnson KW, Januzzi JL, Russak AJ, Paranjpe I, Richter F, et al. Prevalence and Impact of Myocardial Injury in Patients Hospitalized With COVID-19 Infection. J Am Coll Cardiol. 2020;76(5):533–46.
Zaninotto M, Mion MM, Padoan A, Babuin L, Plebani M. Cardiac troponin I in SARS-CoV-2-patients: The additional prognostic value of serial monitoring. Clin Chim Acta. 2020;511:75–80.
Fujino M, Ishii M, Taniguchi T, Chiba H, Kimata M, Hitosugi M. The Value of Interleukin-6 among Several Inflammatory Markers as a Predictor of Respiratory Failure in COVID-19 Patients. Diagnostics 2021, Vol 11, Page 1327 [Internet]. 2021 Jul 23;11(8):1327. Available from: https://www.mdpi.com/2075-4418/11/8/1327/htm.
Malik P, Patel U, Mehta D, Patel N, Kelkar R, Akrmah M, et al. Biomarkers and outcomes of COVID-19 hospitalisations: systematic review and meta-analysis. BMJ Evidence-Based Med [Internet]. 2021;26(3):107–8 Available from: https://ebm.bmj.com/content/26/3/107.
Akbar MR, Pranata R, Wibowo A, Lim MA, Sihite TA, Martha JW. The prognostic value of elevated creatine kinase to predict poor outcome in patients with COVID-19 - A systematic review and meta-analysis. Diabetes Metab Syndr [Internet]. 2021;15(2):529 Available from: /pmc/articles/PMC7877870/.
De Rosa A, Verrengia EP, Merlo I, Rea F, Siciliano G, Corrao G, et al. Muscle manifestations and CK levels in COVID infection: results of a large cohort of patients inside a Pandemic COVID-19 Area. Acta Myol [Internet]. 2021;40(1):1 Available from: /pmc/articles/PMC8033429/.
Orsucci D, Trezzi M, Anichini R, Blanc P, Barontini L, Biagini C, et al. Increased Creatine Kinase May Predict A Worse COVID-19 Outcome. J Clin Med 2021, Vol 10, Page 1734 [Internet]. 2021 Apr 16;10(8):1734. Available from: https://www.mdpi.com/2077-0383/10/8/1734/htm.
Visacri MB, Nicoletti AS, Pincinato EC, Loren P, Saavedra N, Saavedra K, et al. Role of miRNAs as biomarkers of COVID-19: a scoping review of the status and future directions for research in this field. Biomark Med [Internet]. 2021;15(18):1785–95 Available from: https://www.futuremedicine.com/doi/abs/10.2217/bmm-2021-0348.
Aschenbrenner AC, Mouktaroudi M, Krämer B, Oestreich M, Antonakos N, Nuesch-Germano M, et al. Disease severity-specific neutrophil signatures in blood transcriptomes stratify COVID-19 patients. Genome Med [Internet]. 2021;13(1):7 Available from: https://genomemedicine.biomedcentral.com/articles/10.1186/s13073-020-00823-5.
Rastrelli G, Di Stasi V, Inglese F, Beccaria M, Garuti M, Di Costanzo D, et al. Low testosterone levels predict clinical adverse outcomes in SARS-CoV-2 pneumonia patients. Andrology [Internet]. 2021;9(1):88–98 Available from: https://onlinelibrary.wiley.com/doi/full/10.1111/andr.12821.
He Y, Wang J, Li F, Shi Y. Main Clinical Features of COVID-19 and Potential Prognostic and Therapeutic Value of the Microbiota in SARS-CoV-2 Infections. Front Microbiol. 2020;11:1302.
FDA. Precision Medicine | FDA [Internet]. Available from: https://www.fda.gov/medical-devices/in-vitro-diagnostics/precision-medicine.
Campuzano S, Yánez-Sedeño P, Pingarrón JM. Electrochemical bioaffinity sensors for salivary biomarkers detection. TrAC Trends Anal Chem. 2017;86:14–24.
Verdecchia P, Cavallini C, Spanevello A, Angeli F. The pivotal link between ACE2 deficiency and SARS-CoV-2 infection. Eur J Intern Med. 2020;76:14–20.
Genomewide Association Study of Severe Covid-19 with Respiratory Failure. N Engl J Med [Internet]. 2020;383(16):1522–34 Available from: https://www.nejm.org/doi/full/10.1056/nejmoa2020283.
Drucker DJ. Diabetes, obesity, metabolism, and SARS-CoV-2 infection: the end of the beginning. Cell Metab. 2021;33(3):479–98.
Allegra A, Tonacci A, Pioggia G, Musolino C, Gangemi S. Vitamin deficiency as risk factor for SARS-CoV-2 infection: Correlation with susceptibility and prognosis. Eur Rev Med Pharmacol Sci. 2020;24(18):9721–38.
Hernández JL, Nan D, Fernandez-Ayala M, Garciá-Unzueta M, Hernández-Hernández MA, López-Hoyos M, et al. Vitamin D Status in Hospitalized Patients with SARS-CoV-2 Infection. J Clin Endocrinol Metab [Internet]. 2021;106(3):E1343–53 Available from: /pmc/articles/PMC7797757/.
Toledano JM;, Moreno-Fernandez J;, Puche-Juarez M;, Ochoa JJ;, Diaz-Castro J, Toledano JM, et al. Implications of Vitamins in COVID-19 Prevention and Treatment through Immunomodulatory and Anti-Oxidative Mechanisms. Antioxidants 2022, Vol 11, Page 5 [Internet]. 2021 Dec 21;11(1):5. Available from: https://www.mdpi.com/2076-3921/11/1/5/htm.
Ovsyannikova IG, Haralambieva IH, Crooke SN, Poland GA, Kennedy RB. The role of host genetics in the immune response to SARS-CoV-2 and COVID-19 susceptibility and severity. Immunol Rev [Internet]. 2020;296(1):205–19 Available from: https://onlinelibrary.wiley.com/doi/full/10.1111/imr.12897.
Hou Y, Zhao J, Martin W, Kallianpur A, Chung MK, Jehi L, et al. New insights into genetic susceptibility of COVID-19: An ACE2 and TMPRSS2 polymorphism analysis. BMC Med [Internet]. 2020;18(1):1–8 Available from: https://link.springer.com/articles/10.1186/s12916-020-01673-z.
Kaidashev I, Shlykova O, Izmailova O, Torubara O, Yushchenko Y, Tyshkovska T, et al. Host gene variability and SARS-CoV-2 infection: A review article. Heliyon. 2021;7(8):e07863.
Anastassopoulou C, Gkizarioti Z, Patrinos GP, Tsakris A. Human genetic factors associated with susceptibility to SARS-CoV-2 infection and COVID-19 disease severity. Hum Genomics [Internet]. 2020;14(1):1–8 Available from: https://humgenomics.biomedcentral.com/articles/10.1186/s40246-020-00290-4.
Kumari V, Rastogi S, Sharma V. Emerging Trends in Nanobiosensor. Cham: Springer; 2019. p. 419–47. Available from: https://link.springer.com/chapter/10.1007/978-3-030-17061-5_18
Debnath N, Das S. Nanobiosensor: Current trends and applications. In: NanoBioMedicine [Internet]. Springer Singapore; 2020. p. 389–409. Available from: https://link.springer.com/chapter/10.1007/978-981-32-9898-9_16.
Nguyen HH, Lee SH, Lee UJ, Fermin CD, Kim M. Immobilized enzymes in biosensor applications [Internet]. Vol. 12, Materials. MDPI AG; 2019. p. 121. Available from: www.mdpi.com/journal/materials.
Patel S, Nanda R, Sahoo S, Mohapatra E. Biosensors in health care: The milestones achieved in their development towards lab-on-chip-analysis. Vol. 2016, Biochemistry Research International. Hindawi Publishing Corporation; 2016.
Saxena SK, Khurana SMP, editors. NanoBioMedicine [Internet]. NanoBioMedicine. Singapore: Springer Singapore; 2020. Available from: http://link.springer.com/10.1007/978-981-32-9898-9.
Wongkaew N, Simsek M, Griesche C, Baeumner AJ. Functional Nanomaterials and Nanostructures Enhancing Electrochemical Biosensors and Lab-on-a-Chip Performances: Recent Progress, Applications, and Future Perspective. Chem Rev. 2019;119(1):120–94.
Bhalla N, Jolly P, Formisano N, Estrela P. Introduction to biosensors. Essays Biochem [Internet]. 2016;60(1):1 Available from: /pmc/articles/PMC4986445/.
Sabu C, Henna TK, Raphey VR, Nivitha KP, Pramod K. Advanced biosensors for glucose and insulin. Biosensors and Bioelectronics Elsevier Ltd. 2019;141:111201.
Palestino G, García-Silva I, González-Ortega O, Rosales-Mendoza S. Can nanotechnology help in the fight against COVID-19? Expert Rev Anti Infect Ther [Internet]. 2020;18(9):849–64 Available from: https://www.tandfonline.com/doi/full/10.1080/14787210.2020.1776115.
Sharifi M, Avadi MR, Attar F, Dashtestani F, Ghorchian H, Rezayat SM, et al. Cancer diagnosis using nanomaterials based electrochemical nanobiosensors. Vol. 126, Biosensors and Bioelectronics. Elsevier Ltd; 2019. p. 773–84.
Quinchia J, Echeverri D, Cruz-Pacheco AF, Maldonado ME, Orozco JA. Electrochemical biosensors for determination of colorectal tumor biomarkers. Micromachines. 2020;11(4):1–46.
Dutta G. Nanobiosensor-based diagnostic system: Transducers and surface materials. In: Nanobiomaterial Engineering: Concepts and Their Applications in Biomedicine and Diagnostics [Internet]. Springer Singapore; 2020. p. 1–13. Available from: https://link.springer.com/chapter/10.1007/978-981-32-9840-8_1.
Shankar SS, Shereema RM, Ramachandran V, Sruthi TV, Kumar VBS, Rakhi RB. Carbon Quantum Dot-Modified Carbon Paste Electrode-Based Sensor for Selective and Sensitive Determination of Adrenaline. ACS Omega [Internet]. 2019;4(4):7903–10 Available from: https://pubs.acs.org/doi/full/10.1021/acsomega.9b00230.
Seo G, Lee G, Kim MJ, Baek SH, Choi M, Ku KB, et al. Rapid Detection of COVID-19 Causative Virus (SARS-CoV-2) in Human Nasopharyngeal Swab Specimens Using Field-Effect Transistor-Based Biosensor. ACS Nano. 2020;14(4):5135–42.
Vadlamani BS, Uppal T, Verma SC, Misra M. Functionalized tio2 nanotube-based electrochemical biosensor for rapid detection of sars-cov-2 [Internet]. Vol. 20, Sensors (Switzerland). MDPI AG; 2020. p. 1–10. Available from: /pmc/articles/PMC7589637/?report=abstract
Fabiani L, Saroglia M, Galatà G, De Santis R, Fillo S, Luca V, et al. Magnetic beads combined with carbon black-based screen-printed electrodes for COVID-19: A reliable and miniaturized electrochemical immunosensor for SARS-CoV-2 detection in saliva. Biosens Bioelectron. 2021;171:112686.
Zhao H, Liu F, Xie W, Zhou TC, OuYang J, Jin L, et al. Ultrasensitive supersandwich-type electrochemical sensor for SARS-CoV-2 from the infected COVID-19 patients using a smartphone. Sensors Actuators, B Chem [Internet]. 2021 Jan 15;327:128899. Available from: /pmc/articles/PMC7489230/?report=abstract.
Idili A, Parolo C, Alvarez-Diduk R, Merkoçi A. Rapid and Efficient Detection of the SARS-CoV-2 Spike Protein Using an Electrochemical Aptamer-Based Sensor. ACS Sensors [Internet]. 2021 Aug 10;acssensors.1c01222. Available from: https://pubs.acs.org/doi/abs/10.1021/acssensors.1c01222.
Perdomo SA, Ortega V, Jaramillo-Botero A, Mancilla N, Mosquera-Delacruz JH, Valencia DP, et al. SenSARS: A Low-Cost Portable Electrochemical System for Ultra-Sensitive, near Real-Time, Diagnostics of SARS-CoV-2 Infections. IEEE Trans Instrum Meas. 2021;70.
Vezza VJ, Butterworth A, Lasserre P, Blair EO, MacDonald A, Hannah S, et al. An electrochemical SARS-CoV-2 biosensor inspired by glucose test strip manufacturing processes. Chem Commun (Camb) [Internet]. 2021 Mar 17; Available from: http://www.ncbi.nlm.nih.gov/pubmed/33729260
Mojsoska B, Larsen S, Olsen DA, Madsen JS, Brandslund I, Alatraktchi FA. Rapid SARS-CoV-2 Detection Using Electrochemical Immunosensor. Sensors [Internet]. 2021;21(2):390 Available from: https://www.mdpi.com/1424-8220/21/2/390.
Yousefi H, Mahmud A, Chang D, Das J, Gomis S, Chen JB, et al. Detection of SARS-CoV-2 Viral Particles Using Direct, Reagent-Free Electrochemical Sensing. J Am Chem Soc [Internet]. 2021;143:1722–7. https://doi.org/10.1021/jacs.0c10810.
Zhao J, Fu Z, Li H, Xiong Y, Cai S, Wang C, et al. Magnet-assisted electrochemical immunosensor based on surface-clean Pd-Au nanosheets for sensitive detection of SARS-CoV-2 spike protein. Electrochim Acta [Internet]. 2022;404:139766 Available from: https://linkinghub.elsevier.com/retrieve/pii/S0013468621020491.
Rahmati Z, Roushani M, Hosseini H, Choobin H. Electrochemical immunosensor with Cu2O nanocube coating for detection of SARS-CoV-2 spike protein. Mikrochim Acta [Internet]. 2021;188(3):105 Available from: http://www.ncbi.nlm.nih.gov/pubmed/33651173.
Lin YH, Han Y, Sharma A, AlGhamdi WS, Liu CH, Chang TH, et al. A Tri-Channel Oxide Transistor Concept for the Rapid Detection of Biomolecules Including the SARS-CoV-2 Spike Protein. Adv Mater [Internet]. 2022;34(3):2104608 Available from: https://onlinelibrary.wiley.com/doi/full/10.1002/adma.202104608.
Ayankojo AG, Boroznjak R, Reut J, Öpik A, Syritski V. Molecularly imprinted polymer based electrochemical sensor for quantitative detection of SARS-CoV-2 spike protein. Sensors Actuators B Chem. 2022;353:131160.
Georgas A, Lampas E, Houhoula DP, Skoufias A, Patsilinakos S, Tsafaridis I, et al. ACE2-based capacitance sensor for rapid native SARS-CoV-2 detection in biological fluids and its correlation with real-time PCR. Biosens Bioelectron. 2022;202:114021.
Nascimento ED, Fonseca WT, de Oliveira TR, de Correia CRSTB, Faça VM, de Morais BP, et al. COVID-19 diagnosis by SARS-CoV-2 Spike protein detection in saliva using an ultrasensitive magneto-assay based on disposable electrochemical sensor. Sensors Actuators B Chem. 2022;353:131128.
Soares JC, Soares AC, Angelim MKSC, Proença-Modena JL, Moraes-Vieira PM, Mattoso LHC, et al. Diagnostics of SARS-CoV-2 infection using electrical impedance spectroscopy with an immunosensor to detect the spike protein. Talanta. 2022;239:123076.
Roberts A, Mahari S, Shahdeo D, Gandhi S. Label-free detection of SARS-CoV-2 Spike S1 antigen triggered by electroactive gold nanoparticles on antibody coated fluorine-doped tin oxide (FTO) electrode. Anal Chim Acta. 2021;1188:339207.
Liv L, Kayabay H. An Electrochemical Biosensing Platform for the SARS-CoV-2 Spike Antibody Detection Based on the Functionalised SARS-CoV-2 Spike Antigen Modified Electrode. ChemistrySelect [Internet]. 2022;7(10):e202200256 Available from: https://onlinelibrary.wiley.com/doi/full/10.1002/slct.202200256.
Durmus C, Balaban Hanoglu S, Harmanci D, Moulahoum H, Tok K, Ghorbanizamani F, et al. Indiscriminate SARS-CoV-2 multivariant detection using magnetic nanoparticle-based electrochemical immunosensing. Talanta. 2022;243:123356.
Kumar THV, Srinivasan S, Krishnan V, Vaidyanathan R, Babu KA, Natarajan S, et al. Peptide-Based Direct Electrochemical Detection of Receptor Binding Domains of SARS-CoV-2 Spike Protein in Pristine Samples. SSRN Electron J [Internet]. 2022 Mar 4; Available from: https://papers.ssrn.com/abstract=4044514.
Tabrizi MA, Acedo P. An Electrochemical Impedance Spectroscopy-Based Aptasensor for the Determination of SARS-CoV-2-RBD Using a Carbon Nanofiber–Gold Nanocomposite Modified Screen-Printed Electrode. Biosensors [Internet]. 2022;12(3):142 Available from: https://www.mdpi.com/2079-6374/12/3/142/htm.
Sari AK, Hartati YW, Gaffar S, Anshori I, Hidayat D, Wiraswati HL. The optimization of an electrochemical aptasensor to detect RBD protein S SARS-CoV-2 as a biomarker of COVID-19 using screen-printed carbon electrode/AuNP. J Electrochem Sci Eng [Internet]. 2022;12(1):219–35 Available from: https://www.pub.iapchem.org/ojs/index.php/JESE/article/view/1206.
Mehmandoust M, Gumus ZP, Soylak M, Erk N. Electrochemical immunosensor for rapid and highly sensitive detection of SARS-CoV-2 antigen in the nasal sample. Talanta. 2022;240:123211.
Amouzadeh Tabrizi M, Fernández-Blázquez JP, Medina DM, Acedo P. An ultrasensitive molecularly imprinted polymer-based electrochemical sensor for the determination of SARS-CoV-2-RBD by using macroporous gold screen-printed electrode. Biosens Bioelectron. 2022;196:113729.
Zamzami MA, Rabbani G, Ahmad A, Basalah AA, Al-Sabban WH, Nate Ahn S, et al. Carbon nanotube field-effect transistor (CNT-FET)-based biosensor for rapid detection of SARS-CoV-2 (COVID-19) surface spike protein S1. Bioelectrochemistry. 2022;143:107982.
Singh NK, Ray P, Carlin AF, Magallanes C, Morgan SC, Laurent LC, et al. Hitting the diagnostic sweet spot: Point-of-care SARS-CoV-2 salivary antigen testing with an off-the-shelf glucometer. Biosens Bioelectron [Internet]. 2021;180:113111 Available from: https://linkinghub.elsevier.com/retrieve/pii/S0956566321001482.
Shao W, Shurin MR, Wheeler SE, He X, Star A. Rapid Detection of SARS-CoV-2 Antigens Using High-Purity Semiconducting Single-Walled Carbon Nanotube-Based Field-Effect Transistors. ACS Appl Mater Interfaces [Internet]. 2021;13:10321–7 Available from: http://www.ncbi.nlm.nih.gov/pubmed/33596036.
Raziq A, Kidakova A, Boroznjak R, Reut J, Öpik A, Syritski V. Development of a portable MIP-based electrochemical sensor for detection of SARS-CoV-2 antigen. Biosens Bioelectron [Internet]. 2021 Apr 15 [cited 2021 Mar 19];178:113029. Available from: /pmc/articles/PMC7826012/.
Qi H, Hu Z, Yang Z, Zhang J, Wu JJ, Cheng C, et al. Capacitive Aptasensor Coupled with Microfluidic Enrichment for Real-Time Detection of Trace SARS-CoV-2 Nucleocapsid Protein. Anal Chem [Internet]. 2021; Available from: https://pubs.acs.org/doi/abs/10.1021/acs.analchem.1c04296.
Białobrzeska W, Ficek M, Dec B, Osella S, Trzaskowski B, Jaramillo-Botero A, et al. Performance of electrochemical immunoassays for clinical diagnostics of SARS-CoV-2 based on selective nucleocapsid N protein detection: Boron-doped diamond, gold and glassy carbon evaluation. Biosens Bioelectron [Internet]. 2022;209:114222 Available from: https://linkinghub.elsevier.com/retrieve/pii/S0956566322002627.
Haghayegh F, Salahandish R, Hassani M, Sanati-Nezhad A. Highly Stable Buffer-Based Zinc Oxide/Reduced Graphene Oxide Nanosurface Chemistry for Rapid Immunosensing of SARS-CoV-2 Antigens. ACS Appl Mater Interfaces [Internet]. 2022;14(8):10844–55 Available from: https://pubs.acs.org/doi/abs/10.1021/acsami.1c24475.
Hryniewicz BM, Volpe J, Bach-Toledo L, Kurpel KC, Deller AE, Soares AL, et al. Development of polypyrrole (nano)structures decorated with gold nanoparticles toward immunosensing for COVID-19 serological diagnosis. Mater Today Chem. 2022;24:100817.
Ali MA, Hu C, Zhang F, Jahan S, Yuan B, Saleh MS, et al. N protein-based ultrasensitive SARS-CoV-2 antibody detection in seconds via 3D nanoprinted, microarchitected array electrodes. J Med Virol [Internet]. 2022;94(5):2067–78 Available from: https://onlinelibrary.wiley.com/doi/full/10.1002/jmv.27591.
Hashemi SA, Golab Behbahan NG, Bahrani S, Mousavi SM, Gholami A, Ramakrishna S, et al. Ultra-sensitive viral glycoprotein detection NanoSystem toward accurate tracing SARS-CoV-2 in biological/non-biological media. Biosens Bioelectron [Internet]. 2021;171:112731 Available from: /pmc/articles/PMC7558249/.
Hussein HA, Kandeil A, Gomaa M, Mohamed El Nashar R, El-Sherbiny IM, Hassan RYA. SARS-CoV-2-Impedimetric Biosensor: Virus-Imprinted Chips for Early and Rapid Diagnosis. ACS Sensors [Internet]. 2021;6(11):4098–107 Available from: https://pubs.acs.org/doi/full/10.1021/acssensors.1c01614.
Alafeef M, Dighe K, Moitra P, Pan D. Rapid, Ultrasensitive, and Quantitative Detection of SARS-CoV-2 Using Antisense Oligonucleotides Directed Electrochemical Biosensor Chip. ACS Nano [Internet]. 2020;14(12):17028–45. https://doi.org/10.1021/acsnano.0c06392.
Farzin L, Sadjadi S, Sheini A, Mohagheghpour E. A nanoscale genosensor for early detection of COVID-19 by voltammetric determination of RNA-dependent RNA polymerase (RdRP) sequence of SARS-CoV-2 virus. Mikrochim Acta [Internet]. 2021;188(4):121 Available from: http://www.ncbi.nlm.nih.gov/pubmed/33694010.
Peng Y, Pan Y, Sun Z, Li J, Yi Y, Yang J, et al. An electrochemical biosensor for sensitive analysis of the SARS-CoV-2 RNA. Biosens Bioelectron. 2021;186:113309.
del Caño R, García-Mendiola T, García-Nieto D, Álvaro R, Luna M, Iniesta HA, et al. Amplification-free detection of SARS-CoV-2 using gold nanotriangles functionalized with oligonucleotides. Microchim Acta [Internet]. 2022;189(4):171 Available from: https://link.springer.com/10.1007/s00604-022-05272-y.
Yoon J, Conley BM, Shin M, Choi J-H, Bektas CK, Choi J-W, et al. Ultrasensitive Electrochemical Detection of Mutated Viral RNAs with Single-Nucleotide Resolution Using a Nanoporous Electrode Array (NPEA). ACS Nano [Internet]. 2022 Apr 1;acsnano.1c10824. Available from: https://pubs.acs.org/doi/full/10.1021/acsnano.1c10824.
Ang WL, Lim RRX, Ambrosi A, Bonanni A. Rapid electrochemical detection of COVID-19 genomic sequence with dual-function graphene nanocolloids based biosensor. FlatChem. 2022;32:100336.
Feng Y, Liu G, La M, Liu L. Colorimetric and Electrochemical Methods for the Detection of SARS-CoV-2 Main Protease by Peptide-Triggered Assembly of Gold Nanoparticles. Mol 2022, Vol 27, Page 615 [Internet]. 2022 Jan 18;27(3):615. Available from: https://www.mdpi.com/1420-3049/27/3/615/htm.
Khayamian MA, Parizi MS, Ghaderinia M, Abadijoo H, Vanaei S, Simaee H, et al. A label-free graphene-based impedimetric biosensor for real-time tracing of the cytokine storm in blood serum; suitable for screening COVID-19 patients. RSC Adv [Internet]. 2021;11(55):34503–15 Available from: https://pubs.rsc.org/en/content/articlehtml/2021/ra/d1ra04298j.
Gulias-Cañizo R, Ruíz-Leyja ED, Sánchez-Huerta V, la Paz LO, de. SARS-CoV-2 and the Eye: A Relationship for a Possible Prognostic Tool in COVID-19 Patients. Clin Ophthalmol [Internet]. 2021;15:707 Available from: /pmc/articles/PMC7917349/.
Balne PK, Au VB, Tong L, Ghosh A, Agrawal M, Connolly J, et al. Bead Based Multiplex Assay for Analysis of Tear Cytokine Profiles. J Vis Exp [Internet] 2017;2017(128). Available from: https://pubmed.ncbi.nlm.nih.gov/29053687/.
Jacob JT, Ham B. Compositional profiling and biomarker identification of the tear film. Ocul Surf [Internet]. 2008;6(4):175–85 Available from: https://pubmed.ncbi.nlm.nih.gov/18827950/.
Park J, Kim M, Kim W, Jo S, Kim W, Kim C, et al. Ultrasensitive detection of 25-hydroxy vitamin D3 in real saliva using sandwich-type electrochemical aptasensor. Sensors Actuators B Chem. 2022;355:131239.
Tertis M, Leva PI, Bogdan D, Suciu M, Graur F, Cristea C. Impedimetric aptasensor for the label-free and selective detection of Interleukin-6 for colorectal cancer screening. Biosens Bioelectron. 2019;137:123–32.
Ortega MA, Fernández-Garibay X, Castaño AG, De Chiara F, Hernández-Albors A, Balaguer-Trias J, et al. Muscle-on-a-chip with an on-site multiplexed biosensing system for: In situ monitoring of secreted IL-6 and TNF-α. Lab Chip [Internet]. 2019;19(15):2568–80 Available from: https://pubs.rsc.org/en/content/articlehtml/2019/lc/c9lc00285e.
Yang L, Fan D, Zhang Y, Ding C, Wu D, Wei Q, et al. Ferritin-Based Electrochemiluminescence Nanosurface Energy Transfer System for Procalcitonin Detection Using HWRGWVC Heptapeptide for Site-Oriented Antibody Immobilization. Anal Chem [Internet]. 2019;91(11):7145–52 Available from: https://pubs.acs.org/doi/abs/10.1021/acs.analchem.9b00325.
Liu J, Quan L, Yu X, Wang L. Quantitative detection of procalcitonin using an electrochemical immunosensor based on MoO3/Au@rGO nanocomposites. Analyst [Internet]. 2019;144(23):6968–74 Available from: https://pubs.rsc.org/en/content/articlehtml/2019/an/c9an01721f.
Tanak AS, Jagannath B, Tamrakar Y, Muthukumar S, Prasad S. Non-faradaic electrochemical impedimetric profiling of procalcitonin and C-reactive protein as a dual marker biosensor for early sepsis detection. Anal Chim Acta X. 2019;3:100029.
Gao Z, Li Y, Zhang C, Zhang S, Jia Y, Dong Y. An enzyme-free immunosensor for sensitive determination of procalcitonin using NiFe PBA nanocubes@TB as the sensing matrix. Anal Chim Acta. 2020;1097:169–75.
Abbas Z, Soomro RA, Kalwar NH, Tunesi M, Willander M, Karakuş S, et al. In Situ Growth of CuWO4 Nanospheres over Graphene Oxide for Photoelectrochemical (PEC) Immunosensing of Clinical Biomarker. Sensors [Internet]. 2019;20(1):148 Available from: https://www.mdpi.com/1424-8220/20/1/148.
Medetalibeyoglu H, Beytur M, Akyıldırım O, Atar N, Yola ML. Validated electrochemical immunosensor for ultra-sensitive procalcitonin detection: Carbon electrode modified with gold nanoparticles functionalized sulfur doped MXene as sensor platform and carboxylated graphitic carbon nitride as signal amplification. Sensors Actuators B Chem. 2020;319:128195.
Qu L, Yang L, Ren Y, Ren X, Fan D, Xu K, et al. A signal-off electrochemical sensing platform based on Fe3S4-Pd and pineal mesoporous bioactive glass for procalcitonin detection. Sensors Actuators B Chem. 2020;320:128324.
Molinero-Fernández Á, López MÁ, Escarpa A. An on-chip microfluidic-based electrochemical magneto-immunoassay for the determination of procalcitonin in plasma obtained from sepsis diagnosed preterm neonates. Analyst [Internet]. 2020;145(14):5004–10 Available from: https://pubs.rsc.org/en/content/articlehtml/2020/an/d0an00624f.
Garg M, Chatterjee M, Sharma AL, Singh S. Label-free approach for electrochemical ferritin sensing using biosurfactant stabilized tungsten disulfide quantum dots. Biosens Bioelectron. 2020;151:111979.
Garg M, Rani R, Sharma AL, Singh S. White graphene quantum dots as electrochemical sensing platform for ferritin. Faraday Discuss [Internet]. 2021;227(0):204–12 Available from: https://pubs.rsc.org/en/content/articlehtml/2021/fd/c9fd00111e.
Boonkaew S, Teengam P, Jampasa S, Rengpipat S, Siangproh W, Chailapakul O. Cost-effective paper-based electrochemical immunosensor using a label-free assay for sensitive detection of ferritin. Analyst [Internet]. 2020;145(14):5019–26 Available from: https://pubs.rsc.org/en/content/articlehtml/2020/an/d0an00564a.
Oshin O, Kireev D, Hlukhova H, Idachaba F, Akinwande D, Atayero A. Graphene-Based Biosensor for Early Detection of Iron Deficiency. Sensors [Internet]. 2020;20(13):3688 Available from: https://www.mdpi.com/1424-8220/20/13/3688.
Garg M, Christensen M, Iles A, Sharma A, Singh S, Pamme N. Microfluidic-Based Electrochemical Immunosensing of Ferritin. Biosensors [Internet]. 2020;10(8):91 Available from: https://www.mdpi.com/2079-6374/10/8/91.
Özcan N, Karaman C, Atar N, Karaman O, Yola ML. A Novel Molecularly Imprinting Biosensor Including Graphene Quantum Dots/Multi-Walled Carbon Nanotubes Composite for Interleukin-6 Detection and Electrochemical Biosensor Validation. ECS J Solid State Sci Technol [Internet]. 2020;9(12):121010 Available from: https://iopscience.iop.org/article/10.1149/2162-8777/abd149.
Aydın EB, Aydın M, Sezgintürk MK. A novel electrochemical immunosensor based on acetylene black/epoxy-substituted-polypyrrole polymer composite for the highly sensitive and selective detection of interleukin 6. Talanta. 2021;222:121596.
Gonçalves M de L, Truta LAN, Sales MGF, Moreira FTC. Electrochemical Point-of Care (PoC) Determination of Interleukin-6 (IL-6) Using a Pyrrole (Py) Molecularly Imprinted Polymer (MIP) on a Carbon-Screen Printed Electrode (C-SPE). https://doi.org/10.1080/0003271920211879108 [Internet]. 2021;54(16):2611–23. Available from: https://www.tandfonline.com/doi/abs/10.1080/00032719.2021.1879108.
Zhang C, Shi D, Li X, Yuan J. Microfluidic electrochemical magnetoimmunosensor for ultrasensitive detection of interleukin-6 based on hybrid of AuNPs and graphene. Talanta. 2022;240:123173.
Pandey GR, Kanagavalli P, Karnam K, Thanigai Arul K, Monisha P, Dong CL, et al. Molybdenum trioxide hybridized kaempferol: double-powered nanosystem for salvaging oxidative stress and electrochemical immunoprobing of interleukin-6. Mater Today Chem [Internet]. 2022;24:100809 Available from: https://linkinghub.elsevier.com/retrieve/pii/S2468519422000386.
Qi M, Huang J, Wei H, Cao C, Feng S, Guo Q, et al. Graphene Oxide Thin Film with Dual Function Integrated into a Nanosandwich Device for in Vivo Monitoring of Interleukin-6. ACS Appl Mater Interfaces [Internet]. 2017;9(48):41659–68 Available from: https://pubs.acs.org/doi/abs/10.1021/acsami.7b10753.
Mansuriya BD, Altintas Z. Enzyme-free electrochemical nano-immunosensor based on graphene quantum dots and gold nanoparticles for cardiac biomarker determination. Nanomaterials. 2021;11(3):1–18.
Yola ML, Atar N. Development of cardiac troponin-I biosensor based on boron nitride quantum dots including molecularly imprinted polymer. Biosens Bioelectron. 2019;1(126):418–24.
Sun D, Luo Z, Lu J, Zhang S, Che T, Chen Z, et al. Electrochemical dual-aptamer-based biosensor for nonenzymatic detection of cardiac troponin I by nanohybrid electrocatalysts labeling combined with DNA nanotetrahedron structure. Biosens Bioelectron. 2019;134:49–56.
Chekin F, Vasilescu A, Jijie R, Singh SK, Kurungot S, Iancu M, et al. Sensitive electrochemical detection of cardiac troponin I in serum and saliva by nitrogen-doped porous reduced graphene oxide electrode. Sensors Actuators B Chem. 2018;262:180–7.
Rodrigues VC, Moraes ML, Soares JC, Soares AC, Sanfelice R, Deffune E, et al. Immunosensors Made with Layer-by-Layer Films on Chitosan/Gold Nanoparticle Matrices to Detect D-Dimer as Biomarker for Venous Thromboembolism. https://doi.org/101246/bcsj20180019 [Internet]. 2018 Mar 6;91(6):891–6 Available from: https://www.journal.csj.jp/doi/abs/10.1246/bcsj.20180019.
Tasić N, Cavalcante L, Deffune E, Góes MS, Paixão TRLC, Gonçalves LM. Probeless and label-free impedimetric biosensing of D-dimer using gold nanoparticles conjugated with dihexadecylphosphate on screen-printed carbon electrodes. Electrochim Acta. 2021;397:139244.
Li S, Jiang Y, Eda S, Wu JJ. Low-cost and desktop-fabricated biosensor for rapid and sensitive detection of circulating D-dimer biomarker. IEEE Sensors J. 2019;19(4):1245–51.
Matysiak-Brynda E, Siekiera I, Królikowska A, Donten M, Nowicka AM. Combination of copolymer film (PPy-PPyCOOH) and magnetic nanoparticles as an electroactive and biocompatible platform for electrochemical purposes. Electrochim Acta. 2018;263:454–64.
Wang X, Tao G, Meng Y. Nanogold hollow microsphere-based electrochemical immunosensor for the detection of ferritin in human serum. Microchim Acta 2009 1671 [Internet]. 2009 Oct 19;167(1):147–52. Available from: https://link.springer.com/article/10.1007/s00604-009-0225-4.
Matysiak-Brynda E, Wagner B, Bystrzejewski M, Grudzinski IP, Nowicka AM. The importance of antibody orientation in the electrochemical detection of ferritin. Biosens Bioelectron. 2018;109:83–9.
Song TT, Wang W, Meng LL, Liu Y, Jia XB, Mao X. Electrochemical detection of human ferritin based on gold nanorod reporter probe and cotton thread immunoassay device. Chin Chem Lett. 2017;28(2):226–30.
Chen C-Y, Lehr J, Chen C-Y, Lehr J. Label-free Selective Detection of Protein Markers in the Picomolar Range via a Convenient Voltammetric Sensing Strategy. Electroanalysis [Internet]. 2021;33(3):563–7 Available from: https://onlinelibrary.wiley.com/doi/full/10.1002/elan.202060308.
Ma Y, Yang J, Yang T, Deng Y, Gu M, Wang M, et al. Electrochemical detection of C-reactive protein using functionalized iridium nanoparticles/graphene oxide as a tag. RSC Adv [Internet]. 2020;10(16):9723–9 Available from: https://pubs.rsc.org/en/content/articlehtml/2020/ra/c9ra10386d.
Pinyorospathum C, Chaiyo S, Sae-ung P, Hoven VP, Damsongsang P, Siangproh W, et al. Disposable paper-based electrochemical sensor using thiol-terminated poly(2-methacryloyloxyethyl phosphorylcholine) for the label-free detection of C-reactive protein. Microchim Acta 2019 1867 [Internet]. 2019 Jun 26;186(7):1–10. Available from: https://link.springer.com/article/10.1007/s00604-019-3559-6.
Balayan S, Chauhan N, Chandra R, Jain U. Electrochemical Based C-Reactive Protein (CRP) Sensing Through Molecularly Imprinted Polymer (MIP) Pore Structure Coupled with Bi-Metallic Tuned Screen-Printed Electrode. 2021; Available from: https://biointerfaceresearch.com/.
Ge XY, Zhang JX, Feng YG, Wang AJ, Mei LP, Feng JJ. Label-free electrochemical biosensor for determination of procalcitonin based on graphene-wrapped Co nanoparticles encapsulated in carbon nanobrushes coupled with AuPtCu nanodendrites. Microchim Acta 2022 1893 [Internet]. 2022;189(3):1–13. Available from: https://link.springer.com/article/10.1007/s00604-022-05179-8.
Fang Y, Hu Q, Yu X, Wang L. Ultrasensitive electrochemical immunosensor for procalcitonin with signal enhancement based on zinc nanoparticles functionalized ordered mesoporous carbon-silica nanocomposites. Sensors Actuators B Chem. 2018;258:238–45.
Ding H, Yang L, Jia H, Fan D, Zhang Y, Sun X, et al. Label-free electrochemical immunosensor with palladium nanoparticles functionalized MoS2/NiCo heterostructures for sensitive procalcitonin detection. Sensors Actuators B Chem. 2020;312:127980.
Li Y, Liu L, Liu X, Ren Y, Xu K, Zhang N, et al. A dual-mode PCT electrochemical immunosensor with CuCo2S4 bimetallic sulfides as enhancer. Biosens Bioelectron. 2020;163:112280.
Boonkaew S, Jang I, Noviana E, Siangproh W, Chailapakul O, Henry CS. Electrochemical paper-based analytical device for multiplexed, point-of-care detection of cardiovascular disease biomarkers. Sensors Actuators B Chem. 2021;330:129336.
Tanak AS, Muthukumar S, Krishnan S, Schully KL, Clark DV, Prasad S. Multiplexed cytokine detection using electrochemical point-of-care sensing device towards rapid sepsis endotyping. Biosens Bioelectron. 2021;171:112726.
Li X, Li X, Li D, Zhao M, Wu H, Shen B, et al. Electrochemical biosensor for ultrasensitive exosomal miRNA analysis by cascade primer exchange reaction and MOF@Pt@MOF nanozyme. Biosens Bioelectron. 2020;168:112554.
Hu F, Zhang W, Zhang J, Zhang Q, Sheng T, Gu Y. An electrochemical biosensor for sensitive detection of microRNAs based on target-recycled non-enzymatic amplification. Sensors Actuators B Chem. 2018;271:15–23.
Sarkar T, Bohidar HB, Solanki PR. Carbon dots-modified chitosan based electrochemical biosensing platform for detection of vitamin D. Int J Biol Macromol. 2018;109:687–97.
Chauhan D, Kumar R, Panda AK, Solanki PR. An efficient electrochemical biosensor for Vitamin-D3 detection based on aspartic acid functionalized gadolinium oxide nanorods. J Mater Res Technol. 2019;8(6):5490–503.
Anusha T, Bhavani KS, Shanmukha Kumar JV, Brahman PK, Hassan RYA. Fabrication of electrochemical immunosensor based on GCN-β-CD/Au nanocomposite for the monitoring of vitamin D deficiency. Bioelectrochemistry. 2022;143:107935.
Magar HS, Brahman PK, Hassan RYA. Disposable impedimetric nano-immunochips for the early and rapid diagnosis of Vitamin-D deficiency. Biosens Bioelectron X. 2022;10:100124.
Kaur A, Kapoor S, Bharti A, Rana S, Chaudhary GR, Prabhakar N. Gold-platinum bimetallic nanoparticles coated 3-(aminopropyl)triethoxysilane (APTES) based electrochemical immunosensor for vitamin D estimation. J Electroanal Chem. 2020;873:114400.
Acknowledgments
The authors acknowledge Ruta N and EPM for hosting the Max Planck Tandem Groups.
Funding
VV and JO are grateful for the financial support provided by MINCIENCIAS, the University of Antioquia, and the Max Planck Society through the Cooperation agreement 566-1, 2014.
Author information
Authors and Affiliations
Contributions
Vásquez Viviana: Conceptualization, literature search and data analysis, writing original draft and graphs. Orozco Jahir: Conceptualization, literature search and data analysis, writing, review and editing, and funding acquisition and project administration.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Published in the topical collection Electrochemical Biosensors – Driving Personalized Medicine with guest editors Susana Campuzano Ruiz and Maria Jesus Lobo-Castañón.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Vásquez, V., Orozco, J. Detection of COVID-19-related biomarkers by electrochemical biosensors and potential for diagnosis, prognosis, and prediction of the course of the disease in the context of personalized medicine. Anal Bioanal Chem 415, 1003–1031 (2023). https://doi.org/10.1007/s00216-022-04237-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-022-04237-7