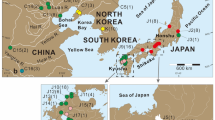
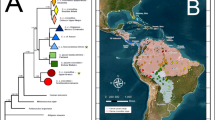
Abstract
Species introductions have been recognized as one of the principal threats to marine environments worldwide. Comparison of genetic data between native and non-native populations can provide key information, such as origin and population demography during the colonization process, which assists in understanding the mechanisms of invasion success in marine environments. The yellowfin goby, Acanthogobius flavimanus, is a large goby native to northeastern Asia, typically inhabiting muddy bottoms of bays, estuaries, and rivers, and is considered a pest where it has invaded coastal areas of the United States and Australia. Here, we analyzed mitochondrial DNA control region sequences of several yellowfin goby populations from both native and non-native distributions. The phylogenetic tree showed no intra-specific lineages, which is in contrast with previous phylogeographic studies that have shown deep genetic divergence in other coastal marine gobies around the Japanese archipelago. On the other hand, at the population level, we found significant genetic differentiation between northern and southern groups in the native distribution, which may be attributed to a rapid population expansion event of the southern group. Our analyses suggest that the origin of the northern California population is Tokyo Bay, but we were unable to identify the original source populations of the southern California and Melbourne populations. These populations showed greatly differing genetic diversities, suggesting their different demographic histories. This study contributes a new perspective on the genetic diversity of multiple populations of the yellowfin goby, as well as representing an example of the relationships between genetic diversity and invasion success.





Similar content being viewed by others
References
Akihito, Sakamoto K, Ikeda Y, Sugiyama K (2002) Suborder Gobioidei. In: Nakabo T (ed) Fishes of Japan with pictorial keys to the species, English edn. Tokai University Press, Tokyo, pp 1139–1310
Akihito, Fumihito A, Ikeda Y et al (2008) Evolution of Pacific Ocean and the Sea of Japan populations of the gobiid species, Pterogobius elapoides and Pterogobius zonoleucus, based on molecular and morphological analyses. Gene 427:7–18
Amsellem L, Noyer J, Le Bourgeois T et al (2000) Comparison of genetic diversity of the invasive weed Rubus alceifolius Poir. (Rosaceae) in its native range and in areas of introduction, using amplified fragment length polymorphism (AFLP) markers. Mol Ecol 9:443–455
Baltz DM (1991) Introduced fishes in marine systems and inland seas. Biol Conserv 56:151–177
Bandelt HJ, Forster P, Röhl A (1999) Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol 16:37–48
Barrett S, Richardson B (1986) Genetic attributes of invading species. In: Groves RH, Burdon JJ (eds) Ecology of biological invasions. Cambridge University Press, Cambridge, pp 21–33
Bell JD, Steffe AS, Talbot RB (1987) The oriental goby, Acanthogobius flavimanus, colonizes a third estuary in New South Wales, Australia. Ichthyol Res 34:227–230
Blaxter J (1986) Development of sense organs and behaviour of teleost larvae with special reference to feeding and predator avoidance. Trans Am Fish Soc 115:98–114
Bohonak AJ (1999) Dispersal, gene flow, and population structure. Q Rev Biol 74:21–45
Bohonak AJ (2002) IBD (isolation by distance): a program for analyses of isolation by distance. J Hered 93:153–154
Bradman H, Grewe P, Appleton B (2011) Direct comparison of mitochondrial markers for the analysis of swordfish population structure. Fish Res 109:95–99
Bray DJ, Gomon MF (2011) Fishes. In: Taxonomic Toolkit for marine life of Port Phillip Bay, Museum Victoria. http://portphillipmarinelife.net.au
Brittan MR, Albrecht AB, Hopkirk JB (1963) An oriental goby collected in the San Joaquin River delta near Stockton, California. Calif Fish Game 49:302–304
Brittan MR, Hopkirk JD, Conners JD et al (1970) Explosive spread of the oriental goby Acanthogobius flavimanus in the San Francisco Bay-Delta region of California. Proc Calif Acad Sci 38:207–214
Brogan MW (1994) Distribution and retention of larval fishes near reefs in the Gulf of California. Mar Ecol Prog Ser 115:1–13
Brown JE, Stepien CA (2009) Invasion genetics of the Eurasian round goby in North America: tracing sources and spread patterns. Mol Ecol 18:64–79
Burton RS (1983) Protein polymorphisms and genetic differentiation of marine invertebrate populations. Mar Biol Lett 4:193–206
Dawson M, Louie K, Barlow M et al (2002) Comparative phylogeography of sympatric sister species, Clevelandia ios and Eucyclogobius newberryi (Teleostei, Gobiidae), across the California Transition Zone. Mol Ecol 11:1065–1075
Dlugosch K, Parker I (2008) Founding events in species invasions: genetic variation, adaptive evolution, and the role of multiple introductions. Mol Ecol 17:431–449
Donaldson KA, Wilson RR Jr (1999) Amphi-panamic geminates of snook (Percoidei: Centropomidae) provide a calibration of the divergence rate in the mitochondrial DNA control region of fishes. Mol Phylogenet Evol 13:208–213
Dotsu Y, Mito S (1955) On the breeding-habits, larvae and young of a goby, Acanthogobius flavimanus (Temminck et Schlegel). Jpn J Ichthyol 4:153–161
Drummond AJ, Rambaut A (2007) BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol 7:214
Dupanloup I, Schneider S, Excoffier L (2002) A simulated annealing approach to define the genetic structure of populations. Mol Ecol 11:2571–2581
Excoffier L, Smouse PE, Quattro JM (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131:479–491
Excoffier L, Laval G, Schneider S (2005) Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol Bioinform Online 1:47–50
Felsenstein J (1993) Phylogeny inference package (PHYLIP). Version 3.5. University of Washington, Seattle
Frankham R, Lees K, Montgomery ME et al (1999) Do population size bottlenecks reduce evolutionary potential? Anim Conserv 2:255–260
Haaker PL (1979) Two Asiatic gobiid fishes, Tridentiger trigonocephalus and Acanthogobius flavimanus, in southern California. Bull South Calif Acad Sci 78:56–61
Hasegawa M, Kishino H, Yano T (1985) Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J Mol Evol 22:160–174
Hirase S, Ikeda M (2014) Divergence of mitochondrial DNA lineage of the rocky intertidal goby Chaenogobius gulosus around the Japanese Archipelago: reference to multiple Pleistocene isolation events in the Sea of Japan. Mar Biol 161:565–574
Hirase S, Ikeda M, Kanno M et al (2012) Phylogeography of the intertidal goby Chaenogobius annularis associated with paleoenvironmental changes around the Japanese Archipelago. Mar Ecol Prog Ser 450:167–179
Hirase S, Takeshima H, Nishida M et al (2016) Parallel mitogenome sequencing alleviates random rooting effect in phylogeography. Genome Biol Evol 8:1267–1278
Hoarau G, Coyer J, Veldsink J et al (2007) Glacial refugia and recolonization pathways in the brown seaweed Fucus serratus. Mol Ecol 16:3606–3616
Hoese D (1973) The introduction of the gobiid fishes Acanthogobius flavimanus and Tridentiger trigonocephalus into Australia. Koolewong 2:3–5
Itaki T, Ikehara K, Motoyama I et al (2004) Abrupt ventilation changes in the Japan Sea over the last 30 ky: evidence from deep-dwelling radiolarians. Palaeogeogr Palaeoclimatol Palaeoecol 208:263–278
Japanese Association of Zoos and Aquariums (2007) Propagation commendation in fiscal year 2006. J Jpn Assoc Zoos Aquar 48:70 (Japan)
Japanese Association of Zoos and Aquariums (2008) Propagation commendation in fiscal year 2007. J Jpn Assoc Zoos Aquar 49:64–65 (Japan)
Jensen JL, Bohonak AJ, Kelley ST (2005) Isolation by distance, web service. BMC Genet 6:13
Kang M, Buckley YM, Lowe AJ (2007) Testing the role of genetic factors across multiple independent invasions of the shrub Scotch broom (Cytisus scoparius). Mol Ecol 16:4662–4673
Kanou K, Sano M, Kohno H (2005) Ontogenetic diet shift, feeding rhythm, and daily ration of juvenile yellowfin goby Acanthogobius flavimanus on a tidal mudflat in the Tama River estuary, central Japan. Ichthyol Res 52:319–324
Katayama S, Sakai K, Iwata T et al (2000) Life history of Japanese common goby Acanthogobius flavimanus in Hiroura Lagoon of Natori River mouth. Bull Miyagi Pref Fish Res Dev Center 16:93–97 (Japan)
Kojima S, Hayashi I, Kim D et al (2004) Phylogeography of an intertidal direct-developing gastropod Batillaria cumingi around the Japanese Islands. Mar Ecol Prog Ser 276:161–172
Kokita T, Nohara K (2011) Phylogeography and historical demography of the anadromous fish Leucopsarion petersii in relation to geological history and oceanography around the Japanese Archipelago. Mol Ecol 20:143–164
Lisiecki LE, Raymo ME (2005) A Pliocene-Pleistocene stack of 57 globally distributed benthic δ18O records. Paleoceanography 20:PA1003
Meng L, Moyle PB, Herbold B (1994) Changes in abundance and distribution of native and introduced fishes of Suisun Marsh. Trans Am Fish Soc 123:498–507
Middleton M (1982) The oriental goby, Acanthogobius flavimanus (Temminck and Schlegel), an introduced fish in the coastal waters of New South Wales, Australia. J Fish Biol 21:513–523
Molnar JL, Gamboa RL, Revenga C et al (2008) Assessing the global threat of invasive species to marine biodiversity. Front Ecol Environ 6:485–492
Neilson ME, Wilson RR (2005) mtDNA singletons as evidence of a post-invasion genetic bottleneck in yellowfin goby Acanthogobius flavimanus from San Francisco Bay, California. Mar Ecol Prog Ser 296:197–208
Ni G, Li Q, Kong L et al (2014) Comparative phylogeography in marginal seas of the northwestern Pacific. Mol Ecol 23:534–548
Polzin T, Daneshmand SV (2003) On Steiner trees and minimum spanning trees in hypergraphs. Oper Res Lett 31:12–20
Prentis PJ, Wilson JR, Dormontt EE et al (2008) Adaptive evolution in invasive species. Trends Plant Sci 13:288–294
Provan J, Wattier RA, Maggs CA (2005) Phylogeographic analysis of the red seaweed Palmaria palmata reveals a Pleistocene marine glacial refugium in the English Channel. Mol Ecol 14:793–803
Rambaut A, Drummond A (2009) Tracer version 1.5. 0. WWW document. http://tree.bio.ed.ac.uk/software/tracer/. Accessed 1 Sept 2016
Rice WR (1989) Analyzing tables of statistical tests. Evolution 43:223–225
Rius M, Turon X, Bernardi G et al (2015) Marine invasion genetics: from spatio-temporal patterns to evolutionary outcomes. Biol Invasions 17:869–885
Rogers AR, Harpending H (1992) Population growth makes waves in the distribution of pairwise genetic differences. Mol Biol Evol 9:552–569
Rousset F (1997) Genetic differentiation and estimation of gene flow from F-statistics under isolation by distance. Genetics 145:1219–1228
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Sakai K, Katayama S, Iwata T (2000) Life history of the Japanese common goby, Acanthogobius flavimanus in the Matsushima Bay. Bull Miyagi Pref Fish Res Dev Center 16:85–92 (Japan)
Shimizu M (1984) Fishes and shellfishes in Tokyo Bay (1). Aquabiology 30:9–13 (Japan)
Slatkin M (1993) Isolation by distance in equilibrium and non-equilibrium populations. Evolution 47:264–279
Suzuki N, Sakurai N, Sugihara T (1989) Development of eggs, larvae and juveniles of the oriental goby Acanthogobius flavimanus reared in the laboratory. Suisan Zoshoku 364:277–289 (Japan)
Tajima F (1989) Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123:585–595
Tamura K (1992) Estimation of the number of nucleotide substitutions when there are strong transition-transversion and G+ C-content biases. Mol Biol Evol 9:678–687
Tamura K, Peterson D, Peterson N et al (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Tringali MD, Bert TM, Seyoum S et al (1999) Molecular phylogenetics and ecological diversification of the transisthmian fish genus Centropomus (Perciformes: Centropomidae). Mol Phylogenet Evol 13:193–207
Villesen P (2007) FaBox: an online toolbox for fasta sequences. Mol Ecol Notes 7:965–968
Vlaming VL (1972) Environmental control of teleost reproductive cycles: a brief review. J Fish Biol 4:131–140
Walsh PS, Metzger DA, Higuchi R (1991) Chelex 100 as a medium for simple extraction of DNA for PCR-based typing from forensic material. Biotechniques 10:506–513
Xia X (2013) DAMBE5: a comprehensive software package for data analysis in molecular biology and evolution. Mol Biol Evol 30:1720–1728
Xia X, Xie Z, Salemi M et al (2003) An index of substitution saturation and its application. Mol Phylogenet Evol 26:1–7
Acknowledgements
The authors thank Kyusyu and Tokushima prefecture lodges of Japan Sport Fishing Foundation, local fishing tackle stores in Japan (Jyosyu-Ya Miyagino store, Point Tokushima store, Anguru Koyaura store, Otaru-fishing PAPA, Kameya-Tsurigu Matsue store), R. Tabata, I. Yokoyama, S. Hayasaka, and T. Mikekado for providing specimens, and S. Matsui, R. Wilson, C. Hayward, M. Lockett, M. McGrouther, and M. Gomon for providing information about yellowfin goby populations. The authors are grateful to the members of the Iwasaki laboratory for helpful comments on this research. This work was supported by the Ministry of Education, Culture, Sports, Science, and Technology (KAKENHI 221S0002 and Project “Construction of the platform for intellectual cooperation”) and the Japan Society for the Promotion of Science (KAKENHI 16H06154 and 26850131). The Australian specimens were collected with support from the Centre for Aquatic Pollution, Identification and Management (CAPIM), Museum Victoria and the Arthur Rylah Institute, Department of Environment, Land, Water and Planning. Additional funding support was received from the Australian Academy of Science (Scientific Visits to Japan, International Linkages Program) and the Australian Society for Fish Biology (Early Career Researcher International Travel Award).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
All applicable international, national, and/or institutional guidelines for the care and use of animals were followed.
Additional information
Responsible Editor: T. Reusch.
Reviewed by undisclosed experts.
Electronic supplementary material
Below is the link to the electronic supplementary material.
227_2017_3137_MOESM1_ESM.eps
Supplementary Fig. 1. Median-joining network of the 137 haplotypes of the yellowfin goby. Each line between the haplotypes indicates a single nucleotide substitution. Small black circles between haplotypes represent intermediate hypothesized haplotypes. Circle sizes reflect the sum of the haplotype frequencies of all locations (EPS 995 kb)
Rights and permissions
About this article
Cite this article
Hirase, S., Chambers, S., Hassell, K. et al. Phylogeography of the yellowfin goby Acanthogobius flavimanus in native and non-native distributions. Mar Biol 164, 106 (2017). https://doi.org/10.1007/s00227-017-3137-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00227-017-3137-6