Abstract
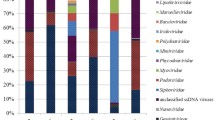
A substantial gap remains in our understanding of the abundance, diversity, and ecology of viruses in soil although some advances have been achieved in recent years. In this study, four soil samples according to the salinity gradient from shore to inland in East China have been characterized. Results showed that spherical virus particles represented the largest viral component in all of the four samples. The viromes had remarkably different taxonomic compositions, and most of the sequences were derived from single-stranded DNA viruses, especially from families Microviridae and Circoviridae. Compared with viromes from other aquatic and sediment samples, the community compositions of our four soil viromes resembled each other, meanwhile coastal sample virome closely congregated with sediment and hypersaline viromes, and high salinity paddy soil sample virome was similar with surface sediment virome. Phylogenetic analysis of functional genes showed that four viromes have high diversity of the subfamily Gokushovirinae in family Microviridae and most of Circoviridae replicase protein sequences grouped within the CRESS-DNA viruses. This work provided an initial outline of the viral communities in marine-terrestrial ecotone and will improve our understanding of the ecological functions of soil viruses.





Similar content being viewed by others
References
Edwards RA, Rohwer F (2005) Viral metagenomics. Nat Rev Microbiol 3:504–510
Chibani-Chennoufi S, Bruttin A, Dillmann ML, Brussow H (2004) Phage-host interaction: an ecological perspective. J Bacteriol 186:3677–3686
Suttle CA (2005) Viruses in the sea. Nature 437:356–361
Dennehy JJ (2014) What ecologists can tell virologists. Annu Rev Microbiol 68:117–135
Wilhelm SW, Suttle CA (1999) Viruses and nutrient cycles in the sea—viruses play critical roles in the structure and function of aquatic food webs. Bioscience 49:781–788
Wommack KE, Colwell RR (2000) Virioplankton: viruses in aquatic ecosystems. Microbiol Mol Biol Rev 64:69–114
Fuhrman JA (1999) Marine viruses and their biogeochemical and ecological effects. Nature 399:541–548
Breitbart M, Salamon P, Andresen B, Mahaffy JM, Segall AM, Mead D, Azam F, Rohwer F (2002) Genomic analysis of uncultured marine viral communities. Proc Natl Acad Sci USA 99:14250–14255
Adriaenssens EM, Van Zyl L, De Maayer P, Rubagotti E, Rybicki E, Tuffin M, Cowan DA (2015) Metagenomic analysis of the viral community in Namib Desert hypoliths. Environ Microbiol 17:480–495
Weinbauer MG (2004) Ecology of prokaryotic viruses. FEMS Microbiol Rev 28:127–181
Weitz JS, Stock CA, Wilhelm SW, Bourouiba L, Coleman ML, Buchan A, Follows MJ, Fuhrman JA, Jover LF, Lennon JT, Middelboe M, Sonderegger DL, Suttle CA, Taylor BP, Thingstad TF, Wilson WH, Wommack KE (2015) A multitrophic model to quantify the effects of marine viruses on microbial food webs and ecosystem processes. ISME J 9:1352–1364
Hurwitz BL, Sullivan MB (2013) The Pacific Ocean Virome (POV): a marine viral metagenomic dataset and associated protein clusters for quantitative viral ecology. PLoS One 8(2):e57355
Mizuno CM, Rodriguez-Valera F, Kimes NE, Ghai R (2013) Expanding the marine virosphere using metagenomics. PLoS Genet 9:e1003987
Yoshida M, Takaki Y, Eitoku M, Nunoura T, Takai K (2013) Metagenomic analysis of viral communities in (hado)pelagic sediments. PLoS One 8(2):e57271
Zablocki O, Adriaenssens EM, Cowan D (2016) Diversity and ecology of viruses in hyperarid desert soils. Appl Environ Microbiol 82:770–777
Reavy B, Swanson MM, Cock PJA, Dawson L, Freitag TE, Singh BK, Torrance L, Mushegian AR, Taliansky M (2015) Distinct circular single-stranded DNA viruses exist in different soil types. Appl Environ Microbiol 81:3934–3945
Hatfull GF, Hendrix RW (2011) Bacteriophages and their genomes. Curr Opin Virol 1:298–303
Hendrix RW (2002) Bacteriophages: evolution of the majority. Theor Popul Biol 61:471–480
Miller RV (2001) Environmental bacteriophage-host interactions: factors contribution to natural transduction. Anton Leeuw Int J G 79:141–147
Williamson KE, Wommack KE, Radosevich M (2003) Sampling natural viral communities from soil for culture-independent analyses. Appl Environ Microbiol 69:6628–6633
Patel A, Noble RT, Steele JA, Schwalbach MS, Hewson I, Fuhrman JA (2007) Virus and prokaryote enumeration from planktonic aquatic environments by epifluorescence microscopy with SYBR Green I. Nat Protoc 2:269–276
Wommack KE, Hill RT, Kessel M, Russekcohen E, Colwell RR (1992) Distribution of viruses in the Chesapeake Bay. Appl Environ Microbiol 58:2965–2970
Zablocki O, van Zyl L, Adriaenssens EM, Rubagotti E, Tuffin M, Cary SC, Cowan D (2014) High-level diversity of tailed phages, eukaryote-associated viruses, and virophage-like elements in the metaviromes of Antarctic soils. Appl Environ Microbiol 80:6888–6897
Dean FB, Nelson JR, Giesler TL, Lasken RS (2001) Rapid amplification of plasmid and phage DNA using phi29 DNA polymerase and multiply-primed rolling circle amplification. Genome Res 11:1095–1099
Wilke A, Bischof J, Gerlach W, Glass E, Harrison T, Keegan KP, Paczian T, Trimble WL, Bagchi S, Gram A, Chaterji S, Meyer F (2016) The MG-RAST metagenomics database and portal in 2015. Nucleic Acids Res 44:D590–D594
Li RQ, Zhu HM, Ruan J, Qian WB, Fang XD, Shi ZB, Li YR, Li ST, Shan G, Kristiansen K, Li SG, Yang HM, Wang J, Wang J (2010) De novo assembly of human genomes with massively parallel short read sequencing. Genome Res 20:265–272
Roux S, Faubladier M, Mahul A, Paulhe N, Bernard A, Debroas D, Enault F (2011) Metavir: a web server dedicated to virome analysis. Bioinformatics 27:3074–3075
Wilke A, Bischof J, Harrison T, Brettin T, D'Souza M, Gerlach W, Matthews H, Paczian T, Wilkening J, Glass EM, Desai N, Meyer F (2015) A RESTful API for accessing microbial community data for MG-RAST. PLoS Comput Biol 11(1):e1004008
Ondov BD, Bergman NH, Phillippy AM (2011) Interactive metagenomic visualization in a Web browser. BMC Bioinf 12:385
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Quaiser A, Dufresne A, Ballaud F, Roux S, Zivanovic Y, Colombet J, Sime-Ngando T, Francez AJ (2015) Diversity and comparative genomics of Microviridae in Sphagnum-dominated peatlands. Front Microbiol 6:375
Rosario K, Dayaram A, Marinov M, Ware J, Kraberger S, Stainton D, Breitbart M, Varsani A (2012) Diverse circular ssDNA viruses discovered in dragonflies (Odonata: Epiprocta). J Gen Virol 93:2668–2681
Williamson KE, Radosevich M, Wommack KE (2005) Abundance and diversity of viruses in six Delaware soils. Appl Environ Microbiol 71:3119–3125
Swanson MM, Fraser G, Daniell TJ, Torrance L, Gregory PJ, Taliansky M (2009) Viruses in soils: morphological diversity and abundance in the rhizosphere. Ann Appl Biol 155:51–60
Wichman HA, Brown CJ (2010) Experimental evolution of viruses: Microviridae as a model system. Philos Trans R Soc B 365:2495–2501
Ackermann HW (2007) 5500 phages examined in the electron microscope. Arch Virol 152:227–243
Ackermann HW (1996) Frequency of morphological phage descriptions in 1995. Arch Virol 141:209–218
Ackermann HW (2001) Frequency of morphological phage descriptions in the year 2000. Arch Virol 146:843–857
Tucker KP, Parsons R, Symonds EM, Breitbart M (2011) Diversity and distribution of single-stranded DNA phages in the North Atlantic Ocean. ISME J 5:822–830
Roux S, Enault F, Robin A, Ravet V, Personnic S, Theil S, Colombet J, Sime-Ngando T, Debroas D (2012) Assessing the diversity and specificity of two freshwater viral communities through metagenomics. PLoS One 7(3):e33641
Labonte JM, Suttle CA (2013) Metagenomic and whole-genome analysis reveals new lineages of gokushoviruses and biogeographic separation in the sea. Front Microbiol 5:52
Roux S, Krupovic M, Poulet A, Debroas D, Enault F (2012) Evolution and diversity of the Microviridae viral family through a collection of 81 new complete genomes assembled from virome reads. PLoS One 7(7):e40418
Zhong X, Guidoni B, Jacas L, Jacquet S (2015) Structure and diversity of ssDNA Microviridae viruses in two peri-alpine lakes (Annecy and Bourget, France). Res Microbiol 166:644–654
Varsani A, Navas-Castillo J, Moriones E, Hernandez-Zepeda C, Idris A, Brown JK, Zerbini FM, Martin DP (2014) Establishment of three new genera in the family Geminiviridae: Becurtovirus, Eragrovirus and Turncurtovirus. Arch Virol 159:2193–2203
Kim KH, Chang HW, Nam YD, Roh SW, Kim MS, Sung Y, Jeon CO, Oh HM, Bae JW (2008) Amplification of uncultured single-stranded DNA viruses from rice paddy soil. Appl Environ Microbiol 74:5975–5985
Cassman N, Prieto-Davo A, Walsh K, Silva GGZ, Angly F, Akhter S, Barott K, Busch J, McDole T, Haggerty JM, Willner D, Alarcon G, Ulloa O, DeLong EF, Dutilh BE, Rohwer F, Dinsdale EA (2012) Oxygen minimum zones harbour novel viral communities with low diversity. Environ Microbiol 14:3043–3065
Guercio A, Savini F, Casa G, Di Marco P, Purpari G, Cannella V, Puleio R, Lavazza A, Buttaci C, Piquemal D, Trentin B, Morent M, Scagliarini A (2014) Massive proliferative cutaneous lesions associated with Poxviridae and Papillomaviridaeviral species in ruminants. Int J Infect Dis 21:183–184
Fierer N, Breitbart M, Nulton J, Salamon P, Lozupone C, Jones R, Robeson M, Edwards RA, Felts B, Rayhawk S, Knight R, Rohwer F, Jackson RB (2007) Metagenomic and small-subunit rRNA analyses reveal the genetic diversity of bacteria, archaea, fungi, and viruses in soil. Appl Environ Microbiol 73:7059–7066
Dinsdale EA, Edwards RA, Hall D, Angly F, Breitbart M, Brulc JM, Furlan M, Desnues C, Haynes M, Li LL, McDaniel L, Moran MA, Nelson KE, Nilsson C, Olson R, Paul J, Brito BR, Ruan YJ, Swan BK, Stevens R, Valentine DL, Thurber RV, Wegley L, White BA, Rohwer F (2008) Functional metagenomic profiling of nine biomes. Nature 452:629–632
Chow CET, Suttle CA (2015) Biogeography of viruses in the sea. Annu Rev Virol 2:41–66
Angly FE, Felts B, Breibart M, Salamon P, Edwards RA, Carlson C, Chan AM, Haynes M, Kelley S, Liu H, Mahaffy JM, Mueller JE, Nulton J, Rohwer F (2006) The marine viromes of four oceanic regions. PLoS Biol 4:2121–2131
Thurber RV, Haynes M, Breitbart M, Wegley L, Rohwer F (2009) Laboratory procedures to generate viral metagenomes. Nat Protoc 4:470–483
Haible D, Kober S, Jeske H (2006) Rolling circle amplification revolutionizes diagnosis and genomics of geminiviruses. J Virol Methods 135:9–16
Kim KH, Bae JW (2011) Amplification methods bias metagenomic libraries of uncultured single-stranded and double-stranded DNA viruses. Appl Environ Microbiol 77:7663–7668
Lizardi PM, Huang XH, Zhu ZR, Bray-Ward P, Thomas DC, Ward DC (1998) Mutation detection and single-molecule counting using isothermal rolling-circle amplification. Nat Genet 19:225–232
Lopez-Bueno A, Tamames J, Velazquez D, Moya A, Quesada A, Alcami A (2009) High diversity of the viral community from an Antarctic Lake. Science 326:858–861
Rosario K, Nilsson C, Lim YW, Ruan YJ, Breitbart M (2009) Metagenomic analysis of viruses in reclaimed water. Environ Microbiol 11:2806–2820
Reyes A, Haynes M, Hanson N, Angly FE, Heath AC, Rohwer F, Gordon JI (2010) Viruses in the faecal microbiota of monozygotic twins and their mothers. Nature 466:334–381
Rodriguez-Brito B, Li LL, Wegley L, Furlan M, Angly F, Breitbart M, Buchanan J, Desnues C, Dinsdale E, Edwards R, Felts B, Haynes M, Liu H, Lipson D, Mahaffy J, Martin-Cuadrado AB, Mira A, Nulton J, Pasic L, Rayhawk S, Rodriguez-Mueller J, Rodriguez-Valera F, Salamon P, Srinagesh S, Thingstad TF, Tran T, Thurber RV, Willner D, Youle M, Rohwer F (2010) Viral and microbial community dynamics in four aquatic environments. ISME J 4:739–751
Fancello L, Trape S, Robert C, Boyer M, Popgeorgiev N, Raoult D, Desnues C (2013) Viruses in the desert: a metagenomic survey of viral communities in four perennial ponds of the Mauritanian Sahara. ISME J 7:359–369
Han LL, Yu DT, Zhang LM, Wang JT, He JZ (2017) Unique community structure of viruses in a glacier soil of the Tianshan Mountains, China. J Soil Sediment 17:852–860
Han LL, Yu DT, Zhang LM, Shen JP, He JZ (2017) Genetic and functional diversity of ubiquitous DNA viruses in selected Chinese agricultural soils. Sci Report 7:45142
Acknowledgments
This work was supported by the National Science Foundation of China (Grant Nos. 41771289 and 41571248).
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Yu, DT., Han, LL., Zhang, LM. et al. Diversity and Distribution Characteristics of Viruses in Soils of a Marine-Terrestrial Ecotone in East China. Microb Ecol 75, 375–386 (2018). https://doi.org/10.1007/s00248-017-1049-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-017-1049-0