Abstract
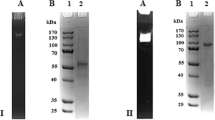
Cellobiose dehydrogenase (CDH) is an extracellular haemoflavoenzyme that is produced by a number of wood-degrading and phytopathogenic fungi and it has a proposed role in the early events of lignocellulose degradation and wood colonisation. In the presence of a suitable electron acceptor, e.g. 2,6-dichloro-indophenol, cytochrome c, or metal ions, CDH oxidises cellobiose to cellobionolactone. When screening 11 different Trametes spp. for the formation of CDH activity, all the strains investigated were found to secrete significant amounts of CDH when cultivated on a cellulose-containing medium. Amongst others, Trametes pubescens and Trametes villosa were identified as excellent, not-yet-described, producer strains of this enzyme activity that has various potential applications in biotechnology. CDH from both strains was purified to apparent homogeneity and subsequently characterised. Both monomeric enzymes have a molecular mass of approximately 90 kDa (gel filtration) and a pI value of 4.2–4.4. The best substrates are cellobiose and cellooligosaccharides; additionally, lactose, thiocellobiose, and xylobiose are efficiently oxidised. Glucose and maltose are poor substrates. The preferred substrate is cellobiose with a K m value of 0.21 mM and a k cat value of 22 s−1 for CDH from T. pubescens; the corresponding values for the T. villosa enzyme are 0.21 mM and 24 s−1, respectively. Both enzymes showed very high activity with one-electron acceptors such as ferricenium, ferricyanide, or the azino-bis-(3-ethyl-benzthiazolin-6-sulfonic acid) cation radical.






Similar content being viewed by others

References
Ansorge W (1985) Fast and sensitive detection of protein and DNA bands by treatment with potassium permanganate. J Biochem Biophys Methods 11:13–20
Baminger U, Nidetzky B, Kulbe KD, Haltrich D (1999) A simple assay for measuring cellobiose dehydrogenase activity in the presence of laccase. J Microbiol Methods 35:253–259
Baminger U, Ludwig R, Galhaup C, Leitner C, Kulbe KD, Haltrich D (2001a) Continuous enzymatic regeneration of redox mediators used in biotransformation reactions employing flavoproteins. J Mol Catal B Enzym 11:541–550
Baminger U, Subramaniam SS, Renganathan V, Haltrich D (2001b) Purification and characterization of cellobiose dehydrogenase from the plant pathogen Sclerotium (Athelia) rolfsii. Appl Environ Microbiol 67:1766–1774
Bao W, Renganathan V (1992) Cellobiose oxidase of Phanerochaete chrysosporium enhances crystalline cellulose degradation by cellulases. FEBS Lett 302:77–80
Bao W, Usha SN, Renganathan V (1993) Purification and characterization of cellobiose dehydrogenase, a novel extracellular hemoflavoenzyme from the white-rot fungus Phanerochaete chrysosporium. Arch Biochem Biophys 300:705–713
Bao W, Lymar E, Renganathan V (1994) Optimization of cellobiose dehydrogenase and β-glucosidase production by cellulose-degrading cultures of Phanerochaete chrysosporium. Appl Microbiol Biotechnol 42:642–646
Cameron MD, Aust SD (2001) Cellobiose dehydrogenase-an extracellular fungal flavocytochrome. Enzyme Microb Technol 28:129–138
Cameron MD, Timofeevski S, Aust SD (2000) Enzymology of Phanerochaete chrysosporium with respect to the degradation of recalcitrant compounds and xenobiotics. Appl Microbiol Biotechnol 54:751–758
Canevascini G, Borer P, Dreyer J-L (1991) Cellobiose dehydrogenases of Sporotrichum (Chrysosporium) thermophile. Eur J Biochem 198:43–52
Daniel G, Volc J, Kubatova E (1994) Pyranose oxidase, a major source of H2O2 during wood degradation by Phanerochaete chrysosporium, Trametes versicolor, and Oudemansiella mucida. Appl Environ Microbiol 60:2524–2532
Dekker RFH (1980) Induction and characterization of a cellobiose dehydrogenase produced by a species of Monilia. J Gen Microbiol 120:309–316
Dumonceaux T, Bartholomew K, Valeanu L, Charles T, Archibald F (2001) Cellobiose dehydrogenase is essential for wood invasion and nonessential for kraft pulp delignification by Trametes versicolor. Enzyme Microb Technol 29:478–489
Elmgren M, Nordling M, Lindquist S-E (1993) The influence of flow rate on biosensors based on redox enzymes incorporated in a redox polymer mounted in a thin-layer flow cells. Anal Biochem 215:261–265
Fang J, Liu W, Gao PJ (1998) Cellobiose dehydrogenase from Schizophyllum commune: purification and study of some catalytic, inactivation, and cellulose-binding properties. Arch Biochem Biophys 353:37–46
Galhaup C, Goller S, Peterbauer CK, Strauss J, Haltrich D (2002) Characterization of the major laccase isoenzyme from Trametes pubescens and regulation of its synthesis by metal ions. Microbiology 148:2159–2169
Habu N, Igarashi K, Samejima M, Pettersson B, Eriksson K-E (1997) Enhanced production of cellobiose dehydrogenase in cultures of Phanerochaete chrysosporium supplemented with bovine calf serum. Biotechnol Appl Biochem 26:97–102
Hai PQ, Nozaki K, Amano Y, Kanda T (2000) Purification and characterization of cellobiose dehydrogenase from Irpex lacteus and its adsorption on cellulose. J Appl Glycosci 47:311–318
Hallberg BM, Bergfors T, Bäckbro K, Pettersson G, Henriksson G, Divne C (2000) A new scaffold for binding haem in the cytochrome domain of the extracellular flavocytochrome cellobiose dehydrogenase. Structure 8:79–88
Hallberg BM, Henriksson G, Pettersson G, Divne C (2002) Crystal structure of the flavoprotein domain of the extracellular flavocytochrome cellobiose dehydrogenase. J Mol Biol 315:421–434
Henriksson G, Sild V, Szabo IJ, Pettersson G, Johansson G (1998) Substrate specificity of cellobiose dehydrogenase from Phanerochaete chrysosporium. Biochim Biophys Acta 1383:48–54
Henriksson G, Johansson G, Pettersson G (2000) A critical review of cellobiose dehydrogenases. J Biotechnol 78:93–113
Hüttermann A, Noelle A (1982) Characterization and regulation of cellobiose dehydrogenase in Fomes annosus. Holzforschung 36:283–286
Kremer SM, Wood PM (1992a) Evidence that cellobiose oxidase from Phanerochaete chrysosporium is primarily an iron(III) reductase: Kinetic comparison with neutrophil NADPH oxidase and yeast flavocytochrome b 2. Eur J Biochem 205:133–138
Kremer SM, Wood PM (1992b) Production of Fenton’s reagent by cellobiose oxidase from cellulolytic cultures of Phanerochaete chrysosporium. Eur J Biochem 208:807–814
Lehman, TC, Thorpe C (1990) Alternate electron acceptors for medium-chain acetyl-Co-A dehydrogenase: use of ferricenium salts. Biochemistry 29:10594–10602
Leitner C, Volc J, Haltrich D (2001) Purification and characterization of pyranose oxidase from the white-rot fungus Trametes multicolor. Appl Environ Microbiol 67:3636–3644
Lindgren A, Stoica L, Ruzgas T, Ciucu A, Gorton L (1999) Development of a cellobiose dehydrogenase modified electrode for amperometric detection of diphenols. Analyst 124:527–532
Ludwig R, Haltrich D (2002) Cellobiose dehydrogenase production by Sclerotium species pathogenic to plants. Lett Appl Microbiol 35:261–266
Ludwig R, Haltrich D (2003) Optimisation of cellobiose dehydrogenase production by the fungus Sclerotium (Athelia) rolfsii. Appl Microbiol Biotechnol 61:32–39
Machida Y, Nakanishi T (1984) Purification and properties of pyranose oxidase from Coriolus versicolor. Agric Biol Chem 48:2463–2470
Morpeth F (1991) Cellobiose oxidoreductases. In: Müller F (ed) Chemistry and biochemistry of flavoenzymes, vol 1, CRC, Boca Raton, Fla. pp 337–348
Moukha SM, Dumonceaux TJ, Record E, Archibald FS (1999) Cloning and analysis of Pycnoporus cinnabarinus cellobiose dehydrogenase. Gene 234:23–33
Roy BP, Dumonceaux T, Koukoulas AA, Archibald FS (1996) Purification and characterization of cellobiose dehydrogenases from the white rot fungus Trametes versicolor. Appl Environ Microbiol 62:4417–4427
Sachslehner A, Haltrich D, Nidetzky B, Kulbe KD (1997) Production of hemicellulose- and cellulose-degrading enzymes by various strains of Sclerotium rolfsii. Appl Biochem Biotechnol 63–65:189–201
Schmidhalter DR, Canevascini G (1993) Isolation and characterization of the cellobiose dehydrogenase from the brown-rot fungus Coniophora puteana (Schum ex Fr.) Karst. Arch Biochem Biophys 300:559–563
Schou C, Christensen MH, Schülein M (1998) Characterization of a cellobiose dehydrogenase from Humicola insolens. Biochem J 330:565–571
Sigoillot C, Lomascolo A, Record E, Robert JL, Asther M, Sigoillot JC (2002) Lignocellulolytic and hemicellulolytic system of Pycnoporus cinnabarinus: isolation and characterization of a cellobiose dehydrogenase and a new xylanase. Enzyme Microb Technol 31:876–883
Subramaniam SS, Nagalla SR, Renganathan V (1999) Cloning and characterization of a thermostable cellobiose dehydrogenase from Sporotrichum thermophile. Arch Biochem Biophys 365:223–230
Yoshida M, Ohira T, Igarashi K, Nagasawa H, Aida K, Hallberg BM, Divne C, Nishino T, Samejima M (2001) Production and characterization of recombinant Phanerochaete chrysosporium cellobiose dehydrogenase in the methylotrophic yeast Pichia pastoris. Biosci Biotech Biochem 65:2050–2057
Yoshida M, Ohira T, Igarashi K, Nagasawa H, Samejima M (2002) Molecular cloning and characterization of a cDNA encoding cellobiose dehydrogenase from the wood-rotting fungus Grifola frondosa. FEMS Microbiol Lett 217:225–230
Acknowledgements
We sincerely thank Hansjörg Prillinger for some of the fungal strains. This work was supported by the Austrian Science Fund (Fonds zur Förderung der wissenschaftlichen Forschung), project FWF P14537-B11.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Ludwig, R., Salamon, A., Varga, J. et al. Characterisation of cellobiose dehydrogenases from the white-rot fungi Trametes pubescens and Trametes villosa . Appl Microbiol Biotechnol 64, 213–222 (2004). https://doi.org/10.1007/s00253-003-1501-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-003-1501-6