Abstract
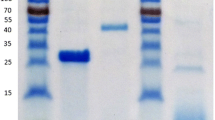
A glycosyl hydrolase family 54 (GH54) α-l-arabinofuranosidase gene (abfA) of Aureobasidium pullulans was amplified by polymerase chain reaction from genomic DNA and a 498-amino-acid open reading frame deduced from the DNA sequence. Modeling of the highly conserved A. pullulans AbfA protein sequence on the crystal structure of Aspergillus kawachii AkabfB showed that the catalytic amino acid arrangement and overall structure were highly similar including the N-terminal catalytic and C-terminal arabinose binding domains. The abfA gene was expressed in Saccharomyces cerevisiae, and the heterologous enzyme was purified. The protein was monomeric, migrating at 49 kDa on sodium dodecyl sulfate-polyacrylamide gel electrophoresis and eluting at 36 kDa upon gel filtration. AbfA showed maximal activity at 55°C and between pH 3.5 and pH 4. The enzyme had a K m value for p-nitrophenyl-α-l-arabinofuranoside of 3.7 mM and a V max of 34.8 μmol min−1 mg protein−1. Arabinose acted as a noncompetitive inhibitor with a K i of 38.4 mM. The enzyme released arabinose from maize fiber, oat spelt arabinoxylan, and wheat arabinoxylan, but not from larch wood arabinogalactan or α-1,5-debranched arabinan. AbfA displayed low activity against α-1,5-l-arabino-oligosaccharides. The enzyme acted synergistically with endo-β-1,4-xylanase in the breakdown of wheat arabinoxylan. Binding of AbfA to xylan from several sources confirmed the presence of a functional carbohydrate-binding module.




Similar content being viewed by others
References
Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K (1995) Current protocols in molecular biology. Wiley, New York
Bacic Q, Harris PJ, Stone BA (1988) Structure and function of plant cell walls. In: Preiss J (ed) Carbohydrates. Academic, San Diego, pp 297–371
De Wet BJM, Van Zyl WH, Prior BA (2006) Characterization of the Aureobasidium pullulans α-glucuronidase expressed in Saccharomyces cerevisiae. Enzyme Microb Technol 38:649–656
Gavel Y, von Heijne G (1990) Sequence differences between glycosylated and non-glycosylated Asn-X-Thr/Ser acceptor sites: implications for protein engineering. Protein Eng 3:433–442
Hill J, Donald KA, Griffiths DE (1991) DMSO-enhanced whole cell yeast transformation. Nucleic Acids Res 19:5791
Jeffries TW (1996) Biochemistry and genetics of microbial xylanases. Curr Opin Biotechnol 7:337–342
Kaneko S, Arimoto M, Ohba M, Kobayashi H, Ishii T, Kusakabe I (1998a) Purification and substrate specificities of two α-l-arabinofuranosidases from Aspergillus awamori IFO 4033. Appl Environ Microbiol 64:4021–4027
Kaneko S, Kuno A, Matsuo N, Ishii T, Kobayashi H, Hayashi K, Kusakabe I (1998b) Substrate specificity of the α-l-arabinofuranosidase from Trichoderma reesei. Biosci Biotechnol Biochem 62:2205–2210
Kormelink FJ, Gruppen H, Voragen AG (1993) Mode of action of (1→4)-β-d-arabinoxylan arabinofuranohydrolase (AXH) and α-l-arabinofuranosidases on alkali-extractable wheat-flour arabinoxylan. Carbohydr Res 249:345–353
Koseki T, Miwa Y, Mese Y, Miyanaga A, Fushinobu S, Wakagi T, Shoun H, Matsuzawa H, Hashizume K (2006) Mutational analysis of N-glycosylation recognition sites on the biochemical properties of Aspergillus kawachii α-l-arabinofuranosidase 54. Biochim Biophys Acta 1760:1458–1464
La Grange DC, Pretorius IS, Van Zyl WH (1996) Expression of a Trichoderma reesei β-xylanase gene (XYN2) in Saccharomyces cerevisiae. Appl Environ Microbiol 62:1036–1044
Leathers TD (1986) Color variants of Aureobasidium pullulans overproduce xylanase with extremely high specific activity. Appl Environ Microbiol 52:1026–1030
Li XL, Ljungdahl LG (1994) Cloning, sequencing, and regulation of a xylanase gene from the fungus Aureobasidium pullulans Y-2311-1. Appl Environ Microbiol 60:3160–3166
Li XL, Ljungdahl LG (1996) Expression of Aureobasidium pullulans xynA in, and secretion of the xylanase from, Saccharomyces cerevisiae. Appl Environ Microbiol 62:209–213
Miyanaga A, Koseki T, Matsuzawa H, Wakagi T, Shoun H, Fushinobu S (2004) Crystal structure of a family 54 α-l-arabinofuranosidase reveals a novel carbohydrate-binding module that can bind arabinose. J Biol Chem 279:44907–44914
Miyanaga A, Koseki T, Miwa Y, Mese Y, Nakamura S, Kuno A, Hirabayashi J, Matsuzawa H, Wakagi T, Shoun H, Fushinobu S (2006) The family 42 carbohydrate-binding module of family 54 α-l-arabinofuranosidase specifically binds the arabinofuranose side chain of hemicellulose. Biochem J 399:503–511
Myburgh J, Prior BA, Kilian SG (1991) The temperature and pH properties of the extracellular hemicellulose-degrading enzymes of Aureobasidium pullulans NRRL Y2311-1. Process Biochem 26:343–348
Nogawa M, Yatsui K, Tomioka A, Okada H, Morikawa Y (1999) An α-l-arabinofuranosidase from Trichoderma reesei containing a noncatalytic xylan-binding domain. Appl Environ Microbiol 65:3964–3968
Ochman H, Gerber AS, Hartl DL (1988) Genetic applications of an inverse polymerase chain reaction. Genetics 120:621–625
Poirot O, O’Toole E, Notredame C (2003) Tcoffee@igs: a web server for computing, evaluating and combining multiple sequence alignments. Nucleic Acids Res 31:3503–3506
Rombouts FM, Voragen AGJ, Searle-van Leeuwen MF, Geraeds CCJM, Schols HA, Pilnik W (1988) The arabinanases of Aspergillus niger—purification and characterisation of two α-l-arabinofuranosidases and an endo-1,5-α-l-arabinanase. Carbohydr Polym 9:25–47
Rose TM, Schultz ER, Henikoff JG, Pietrokovski S, McCallum CM, Henikoff S (1998) Consensus-degenerate hybrid oligonucleotide primers for amplification of distantly related sequences. Nucleic Acids Res 26:1628–1635
Rumbold K, Biely P, Mastihubova M, Gudelj M, Gubitz G, Robra KH, Prior BA (2003) Purification and properties of a feruloyl esterase involved in lignocellulose degradation by Aureobasidium pullulans. Appl Environ Microbiol 69:5622–5626
Saha BC (2000) α-l-Arabinofuranosidases: biochemistry, molecular biology and application in biotechnology. Biotechnol Adv 18:403–423
Saha BC, Bothast RJ (1998) Purification and characterization of a novel thermostable α-l-arabinofuranosidase from a color-variant strain of Aureobasidium pullulans. Appl Environ Microbiol 64:216–220
Sali A, Blundell TL (1993) Comparative protein modeling by satisfaction of spatial restraints. J Mol Biol 234:779–815
Triglia T, Peterson MG, Kemp DJ (1988) A procedure for in vitro amplification of DNA segments that lie outside the boundaries of known sequences. Nucleic Acids Res 16:8186
Von Heijne G (1986) A new method for predicting signal sequence cleavage sites. Nucleic Acids Res 14:4683–4690
Wong KKY, Tan LUL, Saddler JN (1988) Multiplicity of β-1,4-xylanase in microorganisms, functions and applications. Microbiol Rev 52:305–317
Acknowledgment
We thank the National Research Foundation for financial support. The assistance of Marc Claeyssens is gratefully acknowledged with the protein modeling. Excellent technical assistance was provided by Annatjie Hugo and Yolanda Engelbrecht.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Fig. S1
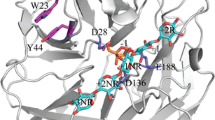
Computer model of the three-dimensional structure of AbfA. a Overlay of the AbfA model (blue) and the solved crystal structure of AkabfB (green). b Molecular surface of AbfA, with the surface colored blue for positive and red for negative according to the electrostatic potential. c Ribbon structure of AbfA, with the four disulfide bridges indicated in yellow. d Topological diagram of the catalytic domains active pocket. The side chains of the amino acid residues, Cys178, Cys179, Met197, Trp208, Asp221, Glu223, Asn224, Leu226, and Asp299, which are important for catalysis, are indicated (GIF 142 kb)
Rights and permissions
About this article
Cite this article
de Wet, B.J.M., Matthew, M.K.A., Storbeck, KH. et al. Characterization of a family 54 α-l-arabinofuranosidase from Aureobasidium pullulans . Appl Microbiol Biotechnol 77, 975–983 (2008). https://doi.org/10.1007/s00253-007-1235-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-007-1235-y