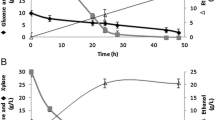
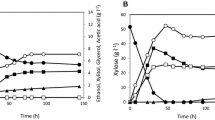
Abstract
Xylitol is industrially synthesized by chemical reduction of d-xylose, which is more expensive than glucose. Thus, there is a growing interest in the production of xylitol from a readily available and much cheaper substrate, such as glucose. The commonly used yeast Pichia pastoris strain GS115 was shown to produce d-arabitol from glucose, and the derivative strain GS225 was obtained to produce twice amount of d-arabitol than GS115 by adaptive evolution during repetitive growth in hyperosmotic medium. We cloned the d-xylulose-forming d-arabitol dehydrogenase (DalD) gene from Klebsiella pneumoniae and the xylitol dehydrogenase (XDH) gene from Gluconobacter oxydans. Recombinant P. pastoris GS225 strains with the DalD gene only or with both DalD and XDH genes could produce xylitol from glucose in a single-fermentation process. Three-liter jar fermentation results showed that recombinant P. pastoris cells with both DalD and XDH converted glucose to xylitol with the highest yield of 0.078 g xylitol/g glucose and productivity of 0.29 g xylitol/L h. This was the first report to convert xylitol from glucose by the pathway of glucose–d-arabitol–d-xylulose–xylitol in a single process. The recombinant yeast could be used as a yeast cell factory and has the potential to produce xylitol from glucose.









Similar content being viewed by others
References
Adachi O, Fujii Y, Ghaly MF, Toyama H, Shinagawa E, Matsushita K (2001) Membrane-bound quinoprotein d-arabitol dehydrogenase of Gluconobacter suboxydans IFO 3257: a versatile enzyme for the oxidative fermentation of various ketoses. Biosci Biotechnol Biochem 65(12):2755–2762
Ahmed Z (2001) The properties of Candida famata R28 for d-arabitol production from d-glucose. Online J Biol Sci 1(11):1005–1008
Akinterinwa O, Cirino PC (2008) Heterologous expression of d-xylulokinase from Pichia stipitis enables high levels of xylitol production by engineered Escherichia coli growing on xylose. Metab Eng 11:48–55
Bae SM, Park YC, Lee TH, Kweon DH, Choi JH, Kim SK, Ryu YW, Seo JH (2004) Production of xylitol by recombinant Saccharomyces cerevisiae containing xylose reductase gene in repeated fed-batch and cell-recycle fermentations. Enzym Microb Technol 35:545–549
Birken S, Pisano MA (1976) Purification and properties of a polyol dehydrogenase from Cephalosporium chrysogenus. J Bacteriol 125:225–232
Blakley ER, Spencer JFT (1962) Study on the formation of d-arabitol by osmophilic yeasts. Can J Biochem Physiol 40:1737–1748
Cheng H, Jiang N (2006) Extremely rapid and efficient DNA extraction from bacteria and yeasts. Biotechnol Lett 28(1):35–39
Cheng H, Jiang N, Shen A, Feng Y (2005) Functional cloning and expression of an NAD-dependent d-arabitol dehydrogenase gene from Gluconobacter oxydans in Escherichia coli. FEMS Microbiol Lett 252(1):35–42
Cheng H, Li Z, Jiang N, Deng Z (2009) Cloning, purification and characterization of an NAD-dependent d-arabitol dehydrogenase from acetic acid bacterium, Acetobacter suboxydans. Protein J 28(6):263–272
Chin JW, Khankal R, Monroe CA, Maranas CD, Cirino PC (2009) Analysis of NADPH supply during xylitol production by engineered Escherichia coli. Biotechnol Bioeng 102(1):209–220
Cirino PC, Chin JW, Ingram LO (2006) Engineering Escherichia coli for xylitol production from glucose–xylose mixtures. Biotechnol Bioeng 95(6):1167–1176
De Schutter K, Lin YC, Tiels P, Van Hecke A, Glinka S, Weber-Lehmann J, Rouzé P, Van de Peer Y, Callewaert N (2009) Genome sequence of the recombinant protein production host Pichia pastoris. Nat Biotechnol 27(6):561–566
Fouts DE, Tyler HL, DeBoy RT, Daugherty S, Ren Q, Badger JH, Durkin AS, Huot H, Shrivastava S, Kothari S, Dodson RJ, Mohamoud Y, Khouri H, Roesch LF, Krogfelt KA, Struve C, Triplett EW, Methé BA (2008) Complete genome sequence of the N2-fixing broad host range endophyte Klebsiella pneumoniae 342 and virulence predictions verified in mice. PLoS Genet 4(7):e1000141
Harkki AM, Myasnikov AN, Apajalahti JHA, Pastinen OA (1997) Manufacturing of xylitol using recombinant microbial hosts. U.S. patent 5,631,150
Harkki AM, Myasnikov AN, Apajalahti JHA, Pastinen OA (2004) Manufacture of xylitol using recombinant microbial hosts. U.S. patent 6,723,540
Huel H, Shakeri-Garakani A, Turgut S, Lengerler JW (1998) Genes for d-arabinitol and ribitol catabolism from Klebsiella pneumoniae. Microbiology 144:1631–1639
Ingram JM, Wood WA (1965) Enzymatic basis for d-arabitol production by Saccharomyces rouxii. J Bacteriol 89:1186–1194
Jin YS, Cruz J, Jeffries TW (2005) Xylitol production by a Pichia stipitis d-xylulokinase mutant. Appl Microbiol Biotechnol 68:42–45
Jovall PA, Tunblad-Johansson I, Adler L (1990) 13C NMR analysis of production and accumulation of osmoregulatory metabolites in the salt-tolerant yeast Debaryomyces hansenii. Arch Microbiol 154:209–214
Ko BS, Kim J, Kim JH (2006) Production of xylitol from d-xylose by a xylitol dehydrogenase gene-disrupted mutant of Candida tropicalis. Appl Environ Microbiol 72(6):4207–4213
Küberl A, Schneider J, Thallinger GG, Anderl I, Wibberg D, Hajek T, Jaenicke S, Brinkrolf K, Goesmann A, Szczepanowski R, Pühler A, Schwab H, Glieder A, Pichler H (2011) High-quality genome sequence of Pichia pastoris CBS7435. J Biotechnol 154:312–320
Lang GI, Rice DP, Hickman MJ, Sodergren E, Weinstock GM, Bostein D, Desai MM (2013) Pervasive genetic hitchhiking and clonal interference in forty evolving yeast populations. Nature. doi:10.1038/nature12344
Li D, Yu J, Tian L, Ji X, Yuan Z (2002) Production of SAM by recombinant Pichia pastoris. Chinese J Biotechnol 18:295–299
Lin Y, Xie Z, Zhang J, Bao W, Pan H, Li B (2012) Cloning and characterization of a novel NAD-dependent xylitol dehydrogenase from Gluconobacter oxydans CGMCC1.637. Acta Microbiol Sin 52:726–735
Lowe DA, Jennings DH (1975) Carbohydrate metabolism in the fungus Dendryphiella salina. V. The pattern of label in arabitol and polysaccharide after growth in the presence of specifically-labelled carbon sources. New Phytol 74:67–79
Maaheimo H, Fiaux J, Cakar ZP, Bailey JE, Sauer U, Szyperski T (2001) Central carbon metabolism of Saccharomyces cerevisiae explored by biosynthetic fractional 13C labeling of common amino acids. Eur J Biochem 268:2464–2479
Mager WH, Siderius M (2002) Novel insights into the osmotic stress response of yeast. FEMS Yeast Res 2:251–257
Mattanovich D, Graf A, Stadlmann J, Dragosits M, Redl A, Maurer M, Kleinheinz M, Sauer M, Altmann F, Gasser B (2009) Genome, secretome and glucose transport highlight unique features of the protein production host Pichia pastoris. Microb Cell Factories 8:29
Meinander NQ, Hahn-Hagerdal B (1997) Influence of cosubstrate concentration on xylose conversion by recombination, XYL1-expressing Saccharomyces cerevisiae : a comparison of different sugars and ethanol as cosubstrate. Appl Environ Microbiol 63:1959–1964
Onishi H, Suzuki T (1969) Microbial production of xylitol from glucose. Appl Microbiol 18:1031–1035
Povelainen M (2008) Pentitol phosphate dehydrogenase: discovery, characterization and use in d-arabitol and xylitol production by metabolically engineered Bacillus subtilis. Dissertation, University of Helsinki
Povelainen M, Miasnikov AN (2007) Production of xylitol by metabolically engineered strains of Bacillus subtilis. J Biotechnol 128:24–31
Pscheidt B, Glierder A (2008) Yeast cell factories for fine chemicals and API production. Microb Cell Factories 7:25
Rao RS, Jyothi Ch P, Prakasham RS, Sarma PN, Rao LV (2006) Xylitol production from corn fiber and sugarcane bagasse hydrolysates by Candida tropicalis. Bioresour Technol 97:1974–1978
Saha BC, Bothast RJ (1999) Production of xylitol by Candida peltata. J Ind Microbiol Biotechnol 22:299–306
Saha BC, Sakakibara Y, Cotta MA (2007) Production of d-arabitol by a newly isolated Zygosaccharomyces rouxii. J Ind Microbiol Biotechnol 34:519–523
Schutter KD, Lin Y-C, Tiels P, van Hecke A, Glinka S, Weber-Lehmann J, Rouzé P, van de Peer Y, Callewaert N (2009) Genome sequence of the recombinant protein production host Pichia pastoris. Nat Biotechnol 27(6):561–566
Silva CJSM, Roberto IC (2001) Improvement of xylitol production by Candida guilliermondii FTI 20037 previously adapted to rice straw hemicellulosic hydrolysate. Lett Appl Microbiol 32(4):248–252
Solà A, Maaheimo H, Ylönen K, Ferrer P, Szyperski T (2004) Amino acid biosynthesis and metabolic flux profiling of Pichia pastoris. Eur J Biochem 271:2462–2470
Steinle A, Witthoff S, Krause JP, Steinbüchel A (2010) Establishment of cyanophycin biosynthesis in Pichia pastoris and optimization by use of engineered cyanophycin synthetases. Appl Environ Microbiol 76:1062–1070
Sugiyama M, Suzuki S, Tonouchi N, Yokozeki K (2003) Cloning of the xylitol dehydrogenase gene from Gluconobacter oxydans and improved production of xylitol from d-arabitol. Biosci Biotechnol Biochem 67(3):584–591
Suzuki S, Sugiyama M, Mihara MY, Hashiguchi K (2002) Novel enzymatic method for the production of xylitol from d-arabitol by Gluconobacter oxydans. Biosci Biotechnol Biochem 66(12):2614–2620
Toivari MH, Ruohonen L, Miasnikov AN, Richard P, Penttila M (2007) Metabolic engineering of Saccharomyces cerevisiae for conversion of d-glucose to xylitol and other five-carbon sugars and sugar alcohols. Appl Environ Microbiol 73(17):5471–5476
Waterham HR, Digan ME, Koutz PJ, Lair SV, Cregg JM (1997) Isolation of the Pichia pastoris glyceraldehyde-3-phosphate dehydrogenase gene and regulation and use of its promoter. Gene 186:37–44
Weimberg R (1962) Mode of formation of d-arabitol by Saccharomyces mellis. Biochem Biophys Res Commun 8:442–445
Wong B, Leeson S, Grindle S, Magee B, Brooks E, Magee PT (1995) d-Arabitol metabolism in Candida albicans: construction and analysis of mutants lacking d-arabitol dehydrogenase. J Bacteriol 177:2971–2976
Acknowledgments
This work was supported by the National High Technology Research and Development Program of China, the National Basic Research Program of China.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Cheng, H., Lv, J., Wang, H. et al. Genetically engineered Pichia pastoris yeast for conversion of glucose to xylitol by a single-fermentation process. Appl Microbiol Biotechnol 98, 3539–3552 (2014). https://doi.org/10.1007/s00253-013-5501-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-013-5501-x