Abstract
Among the different biomarkers predicting response in chronic lymphocytic leukemia (CLL), the most influential parameters are the mutational status of the IGHV genes and the presence of TP53 gene disruptions. Nevertheless, these important assessments are not readily available in most centers dealing with CLL patients. To provide this molecular testing across the country, the Spanish Cooperative Group on CLL (GELLC) established a network of four analytical reference centers. A total of 2153 samples from 256 centers were analyzed over a period of 30 months. In 9% of the patients, we found pathological mutations in the TP53 gene, whereas 48.96% were classified as IGHV unmutated. Results of the satisfaction survey of the program showed a Net Promoter Score of 85.15. Building a national network for molecular testing in CLL allowed the CLL population a broad access to complex biomarkers analysis that should translate into a more accurate and informed therapeutic decision-making.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
During the last 5 years, relevant advances have occurred in the dissection of the genomic, epigenomic, and transcriptomic landscape of chronic lymphocytic leukemia (CLL), the most frequent leukemia in adults [1]. These advances have been translated into further understanding of the prognostic and predictive value of the biological characteristics, particularly genetic lesions associated with chemoresistance (reviewed in [2]).
Among the different genetic lesions described in CLL, deletions of the 17p13.1 chromosomal region (del17p) are found at different frequencies depending on the clinical stage of the disease, ranging from 1% in monoclonal B cell lymphocytosis to 20–40% in chemorefractory cases [3, 4]. Del17p invariably encompasses the locus of the tumor suppressor gene TP53, and mutations of the second allele, usually located in the DNA-binding domain, are found in ~ 80% of cases with del17p. Dysregulation of TP53 is classically linked to refractoriness to chemotherapy and associated with a shorter time to first treatment and overall survival [5,6,7]. More importantly, around 2.5–4.5% of patients requiring front-line therapy harbor TP53 gene mutations without del17p [6, 8,9,10], supporting the notion that in CLL mutational analysis of TP53 gene is complementary to the standard assessment of del17p by fluorescent in situ hybridization (FISH). Accordingly, since TP53 inactivation is determinant in therapeutic decisions in CLL, ascertainment of its disruption is considered mandatory before the onset of any treatment [1].
The mutational status of the IGHV genes represents one of the most important prognostic biomarkers in CLL. Compared with IGHV-mutated (M) cases, CLLs with unmutated IGHV (UM) are characterized by the presence of high-risk genetic lesions, a propensity to undergo clonal evolution, an association with a shorter time to first treatment (TTFT), a shorter progression-free survival when treated with chemotherapy combinations, and poor OS [11,12,13,14,15]. All things considered, the analysis of the IGHV mutational status is highly recommended when assessing the prognosis and determining the therapeutic strategy of patients with CLL [1, 2].
Because of technical difficulties and the need for specifically trained personnel, assessments of mutations of IGHV and TP53 genes are not widely available in the great majority of laboratories dealing with the diagnosis of CLL. This hurdle turned of particular relevance when it became well-established the notion that TP53 dysfunctional cases should not be treated with chemoimmunotherapy, in contrast with what has been observed with novel treatments (reviewed in [2]).
The Spanish Group of CLL (GELLC), with the cooperation of Janssen, launched the TP53 network initiative (RED53), a multicenter task force aimed to facilitate the TP53 and IGHV mutational analysis to the whole country in a due time. The purposes of this network were to provide the mutational analysis of TP53 and IGHV genes to the Spanish centers diagnosing patients with CLL, and to educate hematologists on the need for performing a molecular assessment of CLL patients to guide the therapeutic strategies.
Herein, we report the methodology and general outcomes of building this initiative that enabled the analysis of more than 2000 CLL cases in 2 years, representing another instance of how a national networking can improve the quality of health management of our patients.
Methods
Eligibility of cohort participants
Patients were eligible for the detection of IGHV or TP53 gene mutations when they were diagnosed with CLL and required front-line treatment according to the criteria defined by the IWCLL guidelines [1, 16]. Patients without the need for treatment or those who were previously treated were excluded. Centers from the GELLC that sent samples fulfilled an anonymized form containing the following information of the patient needed for the sample analysis: WBC, lymphocyte count, number of CD19+/CD5+, and lymphocytes. These forms were previously distributed to the centers willing to participate in the network. The program started with the TP53 gene analysis followed by the IGHV testing 1 year later. This project was centrally approved and reviewed by the Ethics Committee from the University Hospital Vall d’Hebron.
Reference centers and procedures
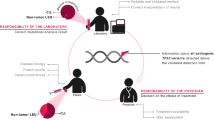
Four Spanish centers (Hospital Clínico, Valencia; Hospital Puerta de Hierro, Madrid; Hospital Universitario, Salamanca; and Hospital Vall d’Hebron, Barcelona) were designated for molecular testing. All the participating centers constituting the network were assigned to send samples to one of those four centers, in such a way that there was an even geographical distribution of centers assigned. These referral centers were certified for TP53 and IGHV analysis according to the criteria determined by the European Initiative for CLL (ERIC) [8, 17].
Physicians willing to perform a genetic test had to collect 8 mL of whole blood in EDTA, obtain an anonymized registry, and send the tube the same day of collection via an express courier to the referral center, adding the form previously mentioned.
The target time for response of TP53 mutational analysis was established in a period inferior to 14 calendar days, and inferior to 21 calendar days for IGHV mutational analysis.
A customer satisfaction survey was performed after 1.5 years of the initiative launching. The goal was to establish the level of service to facilitate benchmarking with a view of enhancing performance. The methodology included to answer an online questionnaire form provided to all the centers participating in the network. In a scale from 1 to 10, items considered were speed of the shipping, quickness of the results, quality of the provided materials and brochures, global satisfaction with the program, degree of recommendation to other professionals, and finally, the Net Promoter Score (NPS) measured according to the standard procedure [18]. Shipping and laboratory testing were supported by Janssen Pharmaceuticals, Spain.
Laboratory methodology
Genomic DNA was extracted from whole blood in EDTA. For IGHV analysis, the clonotypic IGHV-IGHD-IGHJ gene rearrangement was amplified by multiplex PCR using IGHV-leader and IGHJ primers. After Sanger sequencing, the mutational status was determined following ERIC guidelines (Rosenquist et al., Leukemia 2017) [17]. For TP53 mutational analysis, exons 4 to 10 were amplified by PCR followed by Sanger sequencing. Results, interpretation, and reporting were performed following ERIC guidelines [8].
Results
Enrollment of patients
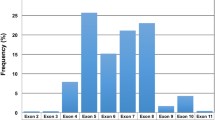
From May 2016 to December 2018, a total of 2153 samples were analyzed from 256 institutions belonging to the Spanish Cooperative Group of CLL (GELLC) (Fig. 1). During the last year, when all the centers included in the study were already participating, the average number of inclusions was 88 patients per month (Fig. 1).
Mutational analysis
Selected referral centers were certified for TP53 and IGHV analysis according to the criteria determined by the European Initiative for CLL (ERIC) [8, 17]. In addition, centers shared samples to ascertain an absolute concordance on the result outputs. For these, samples from four patients with known TP53 somatic mutations were shared among the reference centers. Using the same protocol, all centers classified patients in the same IGHV category and were able to identify and report somatic mutations in TP53.
During this period of analysis, a total of 2153 samples were sent for TP53 gene and/or IGHV mutational analysis. Altogether, 85 samples (3%) had not enough quality for molecular testing, including cases without PCR amplification or with low number of cells.
TP53 gene mutations were tested in 2068 cases. Among them, 182 (9%) patients were classified as having a pathological mutation, and the remaining 1886 patients (91%) were considered wild-type for TP53 (Fig. 2).
As per the IGHV mutational analysis, a total of 1788 patients were analyzed. In 56 cases (3.1%), there was no amplification due to the low number of cells or low quality of samples. Among the remaining 1732 cases, 753 (43.48%) were classified as M-IGHV and the remaining 848 (48.96%) as having UM IGHV genes (Fig. 2). In 7.5% of the cases (n = 131), the result was considered undetermined because of polyclonal rearrangements (n = 34; 1.96%), unproductive (n = 27; 1.56%), or biclonal rearrangements with discordant mutational status (n = 70; 4.04%) were obtained.
Median turnaround time for TP53 mutational status was 9.7 days (95% CI 9.4–9.9 days) which was inferior to the established target time (< 10 days) in 89% of the cases. For the IGHV mutational status analysis, median turnaround time for response was 13 days (95% CI 12.9–14.1 days), again being inferior to the targeted time (< 21 days) in 91% of the cases.
Satisfaction survey
A total of 128 centers (50%) answered the satisfaction survey. Overall assessment of the program was considered excellent. Thus, with a scale from 1 to 10 (being 10 the best), the speed of the shipping was rated 9; quickness on delivering results, 8.65; quality and contents of informative materials and brochures, 9.1 and 9.07, respectively; adhesion to the program, 9.3; and overall satisfaction, 9.14. Finally, the NPS of the project was quantified as 85.15.
Discussion
The most relevant biomarkers in guiding treatment decisions in CLL are the mutational status of IGHV genes and the disruptions of TP53 gene (reviewed in [2]). Unfortunately, in clinical practice limited laboratory capacity is, in general, a bottleneck for these determinations as they require a degree of complexity not always existing in the great majority of centers dealing with patients diagnosed with CLL. To make these determinations accessible to all the patients in the country, the GELLC designed a network of referral centers for IGHV and TP53 mutational testing for CLL patients requiring front-line treatment.
Herein, we present the results obtained during 2 years since the beginning of the network. Only 3.1% of samples arrived to the referral centers in bad condition, which seems acceptable, given the big number of participating centers spread around the country. The results found were consistent with the ones observed in previously reported series of patients having an advanced disease, with a higher percentage of UM IGHV over mutated, and around 10% of cases with pathological gene mutations of TP53. These figures are also resembling the ones observed in front-line clinical trials with chemoimmunotherapy or target treatments [14, 19, 20]. As per IGHV mutations, 7% of cases were considered undetermined. These figures differ from what it was previously reported by ERIC consortium, as they found 11.5% difficult cases and 1.7% cases classified as undetermined [17, 21]. Such differences that could be explained by a variety of reasons: first, at least 30% cases were analyzed using only genomic DNA; second, some of the undetermined cases had not enough tumoral lymphocytes (i.e., small lymphocytic lymphomas). The network will solve part of the undetermined by promoting the use of cDNA in problematic cases and requiring a minimum of 5000 × 109/L malignant lymphocytes to process the sample.
The establishment of this national RED53 network provided several benefits for both patients and physicians, as reflected by the high score observed in the satisfaction survey. First, it facilitated the access to test these important predictive factors to almost all patients requiring therapy. Second, the program educated physicians on the need for assessing molecular biomarkers in patients diagnosed with CLL in need of treatment. Third, the program allowed to build a biobank with part of the samples that should be the basis for future translational research initiatives. Finally, the program enables expertise to be shared and synergies to be brought into play.
A network is an agreement among multiple organizations to address problems which could not be resolved by individual organizations [22]. Networking has become more significant in the health sector for the purpose of advancing the field of science, covering health needs, and prioritizing the allocation of health resources [23]. Laboratory networks are vital to well-functioning public health systems for disease study and control efforts [24,25,26]. As RED53 network organized activities among different groups to increase efficiency, it could be considered a coordinative network [27]. The network presented here served as platform for increasing the quality of care for patients diagnosed with CLL and provided another example of the way scientific initiatives should be designed. The results obtained are consistent with the description of good performance in a collaborative relationship, based on the effectiveness, support, integral character of the solution, and robustness [28]. Such a good performance is based on trust [28], which relay on previously shared experiences within the GELLC and by the professional reputation of the four centers in charge of the tests. Finally, this network supports the partnership between disease cooperative groups and pharmaceutical companies, and brings clinical practice closer to precision medicine. It can be considered a successful example of public-private partnership due to the nature of the participants and the results provided, which, as identified by several authors, include a better value for money and a reduction of some pressure on public budgets, allowing at the same time for a greater innovation [29].
In conclusion, this coordinative network for molecular testing allowed for an increased quality of care for patients with a cancer diagnosis. We conceive that this model could be translated to other countries or cooperative groups for the benefit of patients with momentous diseases.
References
Hallek M, Cheson BD, Catovsky D, Caligaris-Cappio F, Dighiero G, Döhner H, Hillmen P, Keating M, Montserrat E, Chiorazzi N, Stilgenbauer S, Rai KR, Byrd JC, Eichhorst B, O’Brien S, Robak T, Seymour JF, Kipps TJ (2018) iwCLL guidelines for diagnosis, indications for treatment, response assessment, and supportive management of CLL. Blood 131:2745–2760
Bosch F, Dalla-Favera R (2019) Chronic lymphocytic leukaemia: from genetics to treatment. Nat Rev Clin Oncol 16:684–701
Stilgenbauer S, Schnaiter A, Paschka P, Zenz T, Rossi M, Döhner K, Bühler A, Böttcher S, Ritgen M, Kneba M, Winkler D, Tausch E, Hoth P, Edelmann J, Mertens D, Bullinger L, Bergmann M, Kless S, Mack S, Jäger U, Patten N, Wu L, Wenger MK, Fingerle-Rowson G, Lichter P, Cazzola M, Wendtner CM, Fink AM, Fischer K, Busch R, Hallek M, Döhner H (2014) Gene mutations and treatment outcome in chronic lymphocytic leukemia: results from the CLL8 trial. Blood 123:3247–3254
Zenz T, Eichhorst B, Busch R, Denzel T, Häbe S, Winkler D, Bühler A, Edelmann J, Bergmann M, Hopfinger G, Hensel M, Hallek M, Döhner H, Stilgenbauer S (2010) TP53 mutation and survival in chronic lymphocytic leukemia. J Clin Oncol 28:4473–4479
Döhner H, Stilgenbauer S, Benner A, Leupolt E, Kröber A, Bullinger L, Döhner K, Bentz M, Lichter P (2000) Genomic aberrations and survival in chronic lymphocytic leukemia. N Engl J Med 343:1910–1916
Zenz T, Kröber A, Scherer K, Häbe S, Bühler A, Benner A, Denzel T, Winkler D, Edelmann J, Schwänen C, Döhner H, Stilgenbauer S (2008) Monoallelic TP53 inactivation is associated with poor prognosis in chronic lymphocytic leukemia: results from a detailed genetic characterization with long-term follow-up. Blood 112:3322–3329
Rossi D, Rasi S, Spina V, Bruscaggin A, Monti S, Ciardullo C, Deambrogi C, Khiabanian H, Serra R, Bertoni F, Forconi F, Laurenti L, Marasca R, Dal-Bo M, Rossi FM, Bulian P, Nomdedeu J, del Poeta G, Gattei V, Pasqualucci L, Rabadan R, Foà R, Dalla-Favera R, Gaidano G (2013) Integrated mutational and cytogenetic analysis identifies new prognostic subgroups in chronic lymphocytic leukemia. Blood 121:1403–1412
Malcikova J, Tausch E, Rossi D, Sutton LA, Soussi T, Zenz T, Kater AP, Niemann CU, Gonzalez D, Davi F, Gonzalez Diaz M, Moreno C, Gaidano G, Stamatopoulos K, Rosenquist R, Stilgenbauer S, Ghia P, Pospisilova S, European Research Initiative on Chronic Lymphocytic Leukemia (ERIC) — TP53 network (2018) ERIC recommendations for TP53 mutation analysis in chronic lymphocytic leukemia—update on methodological approaches and results interpretation. Leukemia 32:1070–1080
Zenz T, Vollmer D, Trbusek M et al (2010) TP53 mutation profile in chronic lymphocytic leukemia: evidence for a disease specific profile from a comprehensive analysis of 268 mutations. Leukemia 24:2072–2079
Gonzalez D, Martinez P, Wade R, Hockley S, Oscier D, Matutes E, Dearden CE, Richards SM, Catovsky D, Morgan GJ (2011) Mutational status of the TP53 gene as a predictor of response and survival in patients with chronic lymphocytic leukemia: results from the LRF CLL4 trial. J Clin Oncol Off J Am Soc Clin Oncol 29:2223–2229. https://doi.org/10.1200/JCO.2010.32.0838
Hamblin TJ, Davis Z, Gardiner A, Oscier DG, Stevenson FK (1999) Unmutated Ig VH genes are associated with a more aggressive form of chronic lymphocytic leukemia. Blood 94:1848–1854
Damle RN, Wasil T, Fais F, Ghiotto F, Valetto A, Allen SL, Buchbinder A, Budman D, Dittmar K, Kolitz J, Lichtman SM, Schulman P, Vinciguerra VP, Rai KR, Ferrarini M, Chiorazzi N (1999) Ig V gene mutation status and CD38 expression as novel prognostic indicators in chronic lymphocytic leukemia. Blood 94:1840–1847
Landau DA, Carter SL, Stojanov P, McKenna A, Stevenson K, Lawrence MS, Sougnez C, Stewart C, Sivachenko A, Wang L, Wan Y, Zhang W, Shukla SA, Vartanov A, Fernandes SM, Saksena G, Cibulskis K, Tesar B, Gabriel S, Hacohen N, Meyerson M, Lander ES, Neuberg D, Brown JR, Getz G, Wu CJ (2013) Evolution and impact of subclonal mutations in chronic lymphocytic leukemia. Cell 152:714–726
Fischer K, Bahlo J, Fink AM, Goede V, Herling CD, Cramer P, Langerbeins P, von Tresckow J, Engelke A, Maurer C, Kovacs G, Herling M, Tausch E, Kreuzer KA, Eichhorst B, Böttcher S, Seymour JF, Ghia P, Marlton P, Kneba M, Wendtner CM, Döhner H, Stilgenbauer S, Hallek M (2016) Long-term remissions after FCR chemoimmunotherapy in previously untreated patients with CLL: updated results of the CLL8 trial. Blood 127:208–215
Thompson PA, Tam CS, O’Brien SM et al (2016) Fludarabine, cyclophosphamide, and rituximab treatment achieves long-term disease-free survival in IGHV-mutated chronic lymphocytic leukemia. Blood 127:303–309
Hallek M, Cheson BD, Catovsky D, Caligaris-Cappio F, Dighiero G, Döhner H, Hillmen P, Keating MJ, Montserrat E, Rai KR, Kipps TJ (2008) Guidelines for the diagnosis and treatment of chronic lymphocytic leukemia: a report from the International Workshop on Chronic Lymphocytic Leukemia updating the National Cancer Institute–Working Group 1996 guidelines. Blood 111:5446–5456
ERIC, the European Research Initiative on CLL, Rosenquist R, Ghia P et al (2017) Immunoglobulin gene sequence analysis in chronic lymphocytic leukemia: updated ERIC recommendations. Leukemia 31:1477–1481
Reichheld FF (2003) The one number you need to grow. Harv Bus Rev. https://hbr.org/2003/12/the-one-number-you-need-to-grow
Fischer K, Al-Sawaf O, Bahlo J et al (2019) Venetoclax and obinutuzumab in patients with CLL and coexisting conditions. N Engl J Med 380:2225–2236
Goede V, Fischer K, Busch R, Engelke A, Eichhorst B, Wendtner CM, Chagorova T, de la Serna J, Dilhuydy MS, Illmer T, Opat S, Owen CJ, Samoylova O, Kreuzer KA, Stilgenbauer S, Döhner H, Langerak AW, Ritgen M, Kneba M, Asikanius E, Humphrey K, Wenger M, Hallek M (2014) Obinutuzumab plus chlorambucil in patients with CLL and coexisting conditions. N Engl J Med 370:1101–1110
Langerak AW, Davi F, Ghia P, Hadzidimitriou A, Murray F, Potter KN, Rosenquist R, Stamatopoulos K, Belessi C, European Research Initiative on CLL (ERIC) (2011) Immunoglobulin sequence analysis and prognostication in CLL: guidelines from the ERIC review board for reliable interpretation of problematic cases. Leukemia 25:979–984
Agranoff R, McGuire M (2001) Big questions in public network management research. J Public Adm Res Theory 11:295–326
Torchia M, Calabrò A, Morner M (2015) Public–private partnerships in the health care sector: a systematic review of the literature. Public Manag Rev 17:236–261
Mejía N, Santos F, Claverie-Martín F et al (2013) RenalTube: a network tool for clinical and genetic diagnosis of primary tubulopathies. Eur J Pediatr 172:775–780
Kirk CJ, Shult PA (2010) Developing laboratory networks: a practical guide and application. Public Health Rep 125:102–109
Dean L, Njelesani J, Mulamba C et al (2018) Establishing an international laboratory network for neglected tropical diseases: understanding existing capacity in five WHO regions. F1000Research 7:1464
Mandell M, Keast R (2007) Evaluating network arrangements: toward revised performance measures. Public Perform Manag Rev 30:574–597
Warsen R, Nederhand J, Klijn EH, Grotenbreg S, Koppenjan J (2018) What makes public-private partnerships work? Survey research into the outcomes and the quality of cooperation in PPPs. Public Manag Rev 20:1165–1185
Hodge GA, Greve C (2017) On public–private partnership performance: a contemporary review. Public Works Manag Policy 22:55–78
Acknowledgments
The authors thank the patients for their participation.
Funding
This work was supported in part by Janssen Pharmaceutical Companies of Johnson & Johnson. MC holds a contract from Ministerio de Ciencia, Innovación y Universidades (RYC-2012-12018).
Author information
Authors and Affiliations
Contributions
FB, MT, MG, and JAGM designed the project; all the authors contributed to data generation and analysis. All the authors supervised the work. FB wrote the manuscript. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
FB has received research funding and honoraria from Roche, Celgene, Takeda, AstraZeneca, Novartis, Abbie, and Janssen. MC has received research funding from Karyopharm, Pharmacyclics, Roche, Arqule, and AstraZeneca. PA has received honoraria from Janssen, Roche, Celgene, and Abbie. MG has received research funding from Gilead Sciences and honoraria from AbbVie, Gilead, Janssen, and Roche. MA has received honoraria and/or non-financial support from AbbVie, Janssen, and Roche. MT has received research funding and honoraria from Roche, Celgene, Takeda, AstraZeneca, AbbVie, and Janssen. JAGM has received research funding and honoraria from AbbVie, Gilead, Janssen, and Roche. JBS has received research funding from AbbVie, and LGS has received research funding from Janssen. BN and AS have received honoraria from Janssen.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. The Ethical Committee of the University Hospital Vall d’Hebron considered that all the samples obtained were part of the standard of care of the patients, that data was double-anonymized, and that clinical information was not provided following all the ethical requirements.
Informed consent
This article does not contain any studies with human participants performed by any of the authors. Therefore, they stated that no informed consent from the patient was required for this project.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Bosch, F., Navarro, B., Crespo, M. et al. Building a network of TP53 and IGHV testing reference centers across Spain: the Red53 initiative. Ann Hematol 100, 825–830 (2021). https://doi.org/10.1007/s00277-020-04331-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00277-020-04331-9