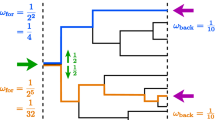
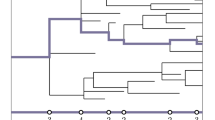
Abstract
Motivated by a recently proposed design for a DNA coded randomised algorithm that enables inference of the average generation of a collection of cells descendent from a common progenitor, here we establish strong convergence properties for the average generation of a super-critical Bellman–Harris process. We further extend those results to a two-type Bellman–Harris process where one type can give rise to the other, but not vice versa. These results further affirm the estimation method’s potential utility by establishing its long run accuracy on individual sample-paths, and significantly expanding its remit to encompass cellular development that gives rise to differentiated offspring with distinct population dynamics.







Similar content being viewed by others
References
Akashi K, Traver D, Miyamoto T, Weissman IL (2000) A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature 404(6774):193
Akinduro O, Weber TS, Ang H, Haltalli MLR, Ruvio N, Duarte D, Rashidi NM, Hawkins ED, Duffy KR, Lo-Celso C (2018) Proliferation dynamics of acute myeloid leukaemia and haematopoietic progenitors competing for bone marrow space. Nat Commun 9:519
Allsopp RC, Vaziri H, Patterson C, Goldstein S, Younglai EV, Futcher AB, Greider CW, Harley CB (1992) Telomere length predicts replicative capacity of human fibroblasts. Proc Natl Acad Sci USA 89(21):10114–10118
Asmussen S (1998) A probabilistic look at the Wiener–Hopf equation. SIAM Rev 40(2):189–201
Asmussen S, Hering H (1983) Branching processes. Birkhauser, Boston
Athreya KB, Jagers P (2012) Classical and modern branching processes. Springer, Berlin
Athreya KB, Ney PE (1972) Branching processes. Courier Corporation, North Chelmsford
Athreya KB, Kaplan N (1976) Convergence of the age distribution in the one-dimensional supercritical age-dependent branching process. Ann Probab 4(1):38–50
Buchholz VR, Flossdorf M, Hensel I, Kretschmer L, Weissbrich B, Gräf P, Verschoor A, Schiemann M, Höfer T, Busch DH (2013) Disparate individual fates compose robust CD8+ T cell immunity. Science 340(6132):630–635
Carlson CA, Kas A, Kirkwood R, Hays LE, Preston BD, Salipante SJ, Horwitz MS (2012) Decoding cell lineage from acquired mutations using arbitrary deep sequencing. Nat Methods 9(1):78–80
Casella G, Berger RL (2002) Statistical inference, vol 2. Duxbury, Pacific Grove
De Boer RJ, Perelson AS (2013) Quantifying T lymphocyte turnover. J Theor Biol 327:45–87
Deenick EK, Hasbold J, Hodgkin PD (1999) Switching to IgG3, IgG2b, and IgA is division linked and independent, revealing a stochastic framework for describing differentiation. J Immunol 163(9):4707–4714
Duarte D, Hawkins ED, Akinduro O, Ang H, De Filippo K, Kong IY, Haltalli M, Ruivo N, Straszkowski L, Vervoort SJ, McLean C, Weber TS, Khorshed R, Pirillo C, Wei A, Ramasamy SK, Kusumbe AP, Duffy K, Adams RH, Purton LE, Carlin LM, Lo Celso C (2018) Inhibition of endosteal vascular niche remodeling rescues hematopoietic stem cell loss in AML. Cell Stem Cell 22(1):64–77
Duffy KR, Subramanian VG (2009) On the impact of correlation between collaterally consanguineous cells on lymphocyte population dynamics. J Math Biol 59(2):255–285
Duffy KR, Wellard CJ, Markham JF, Zhou JHS, Holmberg R, Hawkins ED, Hasbold J, Dowling MR, Hodgkin PD (2012) Activation-induced B cell fates are selected by intracellular stochastic competition. Science 335(6066):338–341
Feller W (1968) An introduction to probability theory and its applications, vol I. Wiley, London
Foudi A, Hochedlinger K, Van Buren D, Schindler JW, Jaenisch R, Carey V, Hock H (2009) Analysis of histone 2B-GFP retention reveals slowly cycling hematopoietic stem cells. Nat Biotechnol 27(1):84–90
Frank SA, Iwasa Y, Nowak MA (2003) Patterns of cell division and the risk of cancer. Genetics 163(4):1527–1532
Giurumescu CA, Kang S, Planchon TA, Betzig E, Bloomekatz J, Yelon D, Cosman P, Chisholm AD (2012) Quantitative semi-automated analysis of morphogenesis with single-cell resolution in complex embryos. Development 139(22):4271–4279
Gomes FL, Zhang G, Carbonell F, Correa JA, Harris WA, Simons BD, Cayouette M (2011) Reconstruction of rat retinal progenitor cell lineages in vitro reveals a surprising degree of stochasticity in cell fate decisions. Development 138(2):227–235
Haccou P, Jagers P, Vatutin VA (2005) Branching processes: variation, growth, and extinction of populations. Cambridge University Press, Cambridge
Harley CB, Futcher AB, Greider CW (1990) Telomeres shorten during ageing of human fibroblasts. Nat Genet 345(6274):458–460
Harris TE (1963) The theory of branching processes. Springe, Berlin
Hawkins ED, Hommel M, Turner ML, Battye FL, Markham JF, Hodgkin PD (2007a) Measuring lymphocyte proliferation, survival and differentiation using CFSE time-series data. Nat Protoc 2(9):2057–2067
Hawkins ED, Turner ML, Dowling MR, van Gend C, Hodgkin PD (2007b) A model of immune regulation as a consequence of randomized lymphocyte division and death times. Proc Natl Acad Sci USA 104:5032–5037
Hawkins ED, Markham JF, McGuinness LP, Hodgkin PD (2009) A single-cell pedigree analysis of alternative stochastic lymphocyte fates. Proc Natl Acad Sci USA 106(32):13457–13462
Hawkins ED, Duarte D, Akinduro O, Khorshed RA, Passaro D, Nowicka M, Straszkowski L, Scott MK, Rothery S, Ruivo N, Foster K, Waibel M, Johnstone RW, Harrison SJ, Westerman DA, Quach H, Gribben J, Robinson MD, Purton LE, Bonnet D, Lo Celso C (2016) T-cell acute leukaemia exhibits dynamic interactions with bone marrow microenvironments. Nature 538(7626):518–522
Hills M, Lücke K, Chavez EA, Eaves CJ, Lansdorp PM (2009) Probing the mitotic history and developmental stage of hematopoietic cells using single telomere length analysis (STELA). Blood 113(23):5765–5775
Hodgkin PD, Lee J-H, Lyons AB (1996) B cell differentiation and isotype switching is related to division cycle number. J Exp Med 184(1):277–281
Hong WK, Bast RC Jr, Hait WN, Kufe DW, Pollock RE, Weichselbaum RR, Holland JF, Frei III E (2010) Holland–Frei cancer medicine, 8th edn. PMPH, New York
Horton MB, Prevedello G, Marchingo JM, Zhou JHS, Duffy KR, Heinzel S, Hodgkin PD (2018) Multiplexed division tracking dyes for proliferation-based clonal lineage tracing. J Immunol 201(3):1097–1103
Jagers P (1969a) The proportions of individuals of different kinds in two-type populations. A branching process problem arising in biology. J Appl Probab 6(2):249–260
Jagers P (1969b) Renewal theory and the almost sure convergence of branching processes. Ark Mat 7(6):495–504
Jagers P (1975) Branching processes with biological applications. Wiley, London
Jagers P (1989) General branching processes as Markov fields. Stoch Process Their Appl 32(2):183–212
Jagers P (1992) Stabilities and instabilities in population dynamics. J Appl Probab 29(4):770–780
Kimmel M, Axelrod DE (2002) Branching processes in biology. Springer, Berlin
Kondo M, Weissman IL, Akashi K (1997) Identification of clonogenic common lymphoid progenitors in mouse bone marrow. Cell 91(5):661–672
Levinson N (1960) Limiting theorems for age-dependent branching processes. Ill J Math 4(1):100–118
Lyons AB (2000) Analysing cell division in vivo and in vitro using flow cytometric measurement of CFSE dye dilution. J Immunol Methods 243(1):147–154
Lyons AB, Parish CR (1994) Determination of lymphocyte division by flow cytometry. J Immunol Methods 171(1):131–137
Marchingo JM, Kan A, Sutherland RM, Duffy KR, Wellard CJ, Belz GT, Lew AM, Dowling MR, Heinzel S, Hodgkin PD (2014) Antigen affinity, costimulation, and cytokine inputs sum linearly to amplify T cell expansion. Science 346(6213):1123–1127
Marchingo JM, Prevedello G, Kan AJ, Heinzel S, Hodgkin PD, Duffy KR (2016) T cell stimuli independently sum to regulate an inherited clonal division fate. Nat Commun 7:13540
Mascré G, Dekoninck S, Drogat B, Youssef KK, Brohée S, Sotiropoulou PA, Simons BD, Blanpain C (2012) Distinct contribution of stem and progenitor cells to epidermal maintenance. Nature 489(7415):257–264
Mendelsohn J, Howley PM, Israel MA, Gray JW, Thompson CB (2015) The molecular basis of cancer. Saunders, Philadelphia
Merlo LMF, Pepper JW, Reid BJ, Maley CC (2006) Cancer as an evolutionary and ecological process. Nat Rev Cancer 6(12):924–935
Mode CJ (1971) Multitype branching processes. Theory and applications. Elsevier, New York
Mukherjea A, Rao M, Suen S (2006) A note on moment generating functions. Stat Probab Lett 76(11):1185–1189
Nordon RE, Ko K-H, Odell R, Schroeder T (2011) Multi-type branching models to describe cell differentiation programs. J Theor Biol 277(1):7–18
Pauklin S, Vallier L (2013) The cell-cycle state of stem cells determines cell fate propensity. Cell 155(1):135–147
Powell EO (1955) Some features of the generation times of individual bacteria. Biometrika 42:16–44
Quah BJC, Parish CR (2012) New and improved methods for measuring lymphocyte proliferation in vitro and in vivo using CFSE-like fluorescent dyes. J Immunol Methods 379(1):1–14
Reizel Y, Chapal-Ilani N, Adar R, Itzkovitz S, Elbaz J, Maruvka YE, Segev E, Shlush LI, Dekel N, Shapiro E (2011) Colon stem cell and crypt dynamics exposed by cell lineage reconstruction. PLoS Genet 7(7):e1002192
Resnick SI (2013) Adventures in stochastic processes. Springer, Berlin
Richards JL, Zacharias AL, Walton T, Burdick JT, Murray JI (2013) A quantitative model of normal caenorhabditis elegans embryogenesis and its disruption after stress. Dev Biol 374(1):12–23
Rudin W (1976) Principles of mathematical analysis, 3rd edn. International series in pure and applied mathematics. McGraw-Hill Book Co., New York
Rufer N, Brümmendorf TH, Kolvraa S, Bischoff C, Christensen K, Wadsworth L, Schulzer M, Lansdorp PM (1999) Telomere fluorescence measurements in granulocytes and T lymphocyte subsets point to a high turnover of hematopoietic stem cells and memory T cells in early childhood. J Exp Med 190(2):157–168
Sakaue-Sawano A, Kurokawa H, Morimura T, Hanyu A, Hama H, Osawa H, Kashiwagi S, Fukami K, Miyata T, Miyoshi H, Imamura T, Ogawa M, Masai H, Miyawaki A (2008) Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell 132(3):487–498
Samuels ML (1971) Distribution of the branching-process population among generations. J Appl Probab 8:655–667
Shibata D, Navidi W, Salovaara R, Li Z-H, Aaltonen LA (1996) Somatic microsatellite mutations as molecular tumor clocks. Nat Med 2(6):676–681
Shibata D, Tavaré S (2006) Counting divisions in a human somatic cell tree. Cell Cycle 5(6):610–614
Smith JA, Martin L (1973) Do cells cycle? Proc Natl Acad Sci USA 70(4):1263–1267
Subramanian VG, Duffy KR, Turner ML, Hodgkin PD (2008) Determining the expected variability of immune responses using the cyton model. J Math Biol 56(6):861–892
Sulston JE, Schierenberg E, White JG, Thomson JN (1983) The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev Biol 100(1):64–119
Tangye SG, Avery DT, Deenick EK, Hodgkin PD (2003) Intrinsic differences in the proliferation of naive and memory human B cells as a mechanism for enhanced secondary immune responses. J Immunol 170(2):686–694
Tomasetti C, Vogelstein B (2015) Variation in cancer risk among tissues can be explained by the number of stem cell divisions. Science 347(6217):78–81
Tsao J-L, Yatabe Y, Salovaara R, Järvinen HJ, Mecklin J-P, Aaltonen LA, Tavaré S, Shibata D (2000) Genetic reconstruction of individual colorectal tumor histories. Proc Natl Acad Sci USA 97(3):1236–1241
Tumbar T, Guasch G, Greco V, Blanpain C, Lowry WE, Rendl M, Fuchs E (2004) Defining the epithelial stem cell niche in skin. Science 303(5656):359–363
Turner M, Hawkins E, Hodgkin PD (2008) Quantitative regulation of B cell division destiny by signal strength. J Immunol 181(1):374–382
Vaziri H, Dragowska W, Allsopp RC, Thomas TE, Harley CB, Lansdorp PM (1994) Evidence for a mitotic clock in human hematopoietic stem cells: loss of telomeric DNA with age. Proc Natl Acad Sci USA 91(21):9857–9860
Wasserstrom A, Frumkin D, Adar R, Itzkovitz S, Stern T, Kaplan S, Shefer G, Shur I, Zangi L, Reizel Y, Harmelin A, Dor Y, Dekel N, Reisner Y, Benayahu D, Tzahor E, Segal E, Shapiro EY (2008) Estimating cell depth from somatic mutations. PLoS Comput Biol 4(5):e1000058
Watson HW, Galton F (1875) On the probability of the extinction of families. J Anthropol Inst Great Br Irel 4:138–144
Weber TS, Perié L, Duffy KR (2016) Inferring average generation via division-linked labeling. J Math Biol 73(2):491–523
Weinrich SL, Pruzan R, Ma L, Ouellette M, Tesmer VM, Holt SE, Bodnar AG, Lichtsteiner S, Kim NW, Trager JB, Taylor RD, Carlos R, Andrews WH, Wright WE, Shay JW, Harley CB, Morin GB (1997) Reconstitution of human telomerase with the template RNA component hTR and the catalytic protein subunit hTRT. Nat Genet 17(4):498–502
Zhang B, Dai M, Li Q-J, Zhuang Y (2013) Tracking proliferative history in lymphocyte development with cre-mediated sister chromatid recombination. PLoS Genet 9(10):e1003887, 10
Acknowledgements
This work was supported by Science Foundation Ireland Grant 12 IP 1263.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Meli, G., Weber, T.S. & Duffy, K.R. Sample path properties of the average generation of a Bellman–Harris process. J. Math. Biol. 79, 673–704 (2019). https://doi.org/10.1007/s00285-019-01373-0
Received:
Revised:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00285-019-01373-0
Keywords
- Bellman–Harris process
- Sample-path properties
- Average generation inference
- Average tree depth inference
- DNA coded algorithm
- Two-type process