Abstract
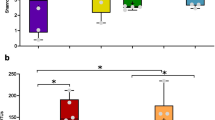
A growing number of studies have provided insights into the diversity of coral-associated bacteria and their function in the coral holobiont. Yet, information about spatial heterogeneity of bacteria within coral colonies is limited. Using 16S rRNA gene metabarcoding, we analyzed the bacterial community composition across four distinct locations in each of five wild Acropora loripes colonies. Considerable variation within and among colonies was present, which has implications for sampling strategies and data interpretation in coral microbiome research. Bacterial assemblages significantly differed in alpha and beta diversity among colonies, with all corals possessing a high relative abundance of Endozoicomonas. When the same A. loripes colonies were subsequently reared in aquaria over 4 weeks, the relative abundance of Marinobacter initially increased in all colonies. However, no significant alteration in bacterial community composition was observed over time and the colonies maintained distinct bacterial microbiomes.





Similar content being viewed by others
Data availability
The sequence data generated and analyzed in this study is available at NCBI under http://www.ncbi.nlm.nih.gov/bioproject/590053, BioProject accession PRJNA590053.
References
Agostini S, Suzuki Y, Higuchi T, Casareto BE, Yoshinaga K, Nakano Y, Fujimura H (2012) Biological and chemical characteristics of the coral gastric cavity. Coral Reefs 31:147–156
Anderson MJ (2001) A new method for non-parametric multivariate analysis of variance. Austral Ecol 26:32–46
Anderson MJ (2006) Distance-based tests for homogeneity of multivariate dispersions. Biometrics 62:245–253
Andersson AF, Lindberg M, Jakobsson H, Bäckhed F, Nyrén P, Engstrand L (2008) Comparative analysis of human gut microbiota by barcoded pyrosequencing. PLoS ONE 3:e2836
Apprill A, Weber L, Santoro AE (2016) Distinguishing between microbial habitats unravels ecological complexity in coral microbiomes. mSystems 1:e00143-16
Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc 57:289–300
Blackall LL, Wilson B, van Oppen MJ (2015) Coral—the world’s most diverse symbiotic ecosystem. Mol Ecol 24:5330–5347
Bokulich NA, Subramanian S, Faith JJ, Gevers D, Gordon JI, Knight R, Mills DA, Caporaso JG (2013) Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat Methods 10:57–59
Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, Alexander H, Alm EJ, Arumugam M, Asnicar F, Bai Y, Bisanz JE, Bittinger K, Brejnrod A, Brislawn CJ, Brown CT, Callahan BJ, Caraballo-Rodríguez AM, Chase J, Cope EK, Silva RD, Diener C, Dorrestein PC, Douglas GM, Durall DM, Duvallet C, Edwardson CF, Ernst M, Estaki M, Fouquier J, Gauglitz JM, Gibbons SM, Gibson DL, Gonzalez A, Gorlick K, Guo J, Hillmann B, Holmes S, Holste H, Huttenhower C, Huttley GA, Janssen S, Jarmusch AK, Jiang L, Kaehler BD, Kang KB, Keefe CR, Keim P, Kelley ST, Knights D, Koester I, Kosciolek T, Kreps J, Langille MGI, Lee J, Ley R, Liu Y-X, Loftfield E, Lozupone C, Maher M, Marotz C, Martin BD, McDonald D, McIver LJ, Melnik AV, Metcalf JL, Morgan SC, Morton JT, Naimey AT, Navas-Molina JA, Nothias LF, Orchanian SB, Pearson T, Peoples SL, Petras D, Preuss ML, Pruesse E, Rasmussen LB, Rivers A, Robeson MS, Rosenthal P, Segata N, Shaffer M, Shiffer A, Sinha R, Song SJ, Spear JR, Swafford AD, Thompson LR, Torres PJ, Trinh P, Tripathi A, Turnbaugh PJ, Ul-Hasan S, van der Hooft JJJ, Vargas F, Vázquez-Baeza Y, Vogtmann E, von Hippel M, Walters W, Wan Y, Wang M, Warren J, Weber KC, Williamson CHD, Willis AD, Xu ZZ, Zaneveld JR, Zhang Y, Zhu Q, Knight R, Caporaso JG (2019) Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol 37:852–857
Bourne DG, Morrow KM, Webster NS (2016) Insights into the coral microbiome: underpinning the health and resilience of reef ecosystems. Annu Rev Microbiol 70:317–340
Callahan BJ, McMurdie PJ, Holmes SP (2017) Exact sequence variants should replace operational taxonomic units in marker-gene data analysis. ISME J 11:2639–2643
Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJ, Holmes SP (2016) DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods 13:581–583
Chan WY, Peplow LM, Menendez P, Hoffmann AA, van Oppen MJH (2019) The roles of age, parentage and environment on bacterial and algal endosymbiont communities in Acropora corals. Mol Ecol 28(16):3830–3843
Collingro A, Tischler P, Weinmaier T, Penz T, Heinz E, Brunham RC, Read TD, Bavoil PM, Sachse K, Kahane S, Friedman MG, Rattei T, Myers GSA, Horn M (2011) Unity in variety—the pan-genome of the chlamydiae. Mol Biol Evol 28:3253–3270
Damjanovic K, Menendez P, Blackall LL, van Oppen MJH (2019) Early life stages of a common broadcast spawning coral associate with specific bacterial communities despite lack of internalized bacteria. Microbial Ecol. https://doi.org/10.1007/s00248-019-01428-1
Damjanovic K, Blackall LL, Menéndez P, van Oppen MJH (2020a) Bacterial and algal symbiont dynamics in early recruits exposed to two adult coral species. Coral Reefs 39:189–202
Damjanovic K, Menéndez P, Blackall LL, van Oppen MJH (2020b) Mixed-mode bacterial transmission in the common brooding coral Pocillopora acuta. Environ Microbiol 22:397–412
Daniels CA, Zeifman A, Heym K, Ritchie KB, Watson CA, Berzins I, Breitbart M (2011) Spatial heterogeneity of bacterial communities in the mucus of Montastraea annularis. Mar Ecol Prog Ser 426:29–40
De Cáceres M, Legendre P (2009) Associations between species and groups of sites: indices and statistical inference. Ecology 90:3566–3574
Epstein H, Torda G, Munday PL, van Oppen MJH (2019) Parental and early life stage environments drive establishment of bacterial and dinoflagellate communities in a common coral. ISME J 13:1635–1638
Glasl B, Smith CE, Bourne DG, Webster NS (2019) Disentangling the effect of host-genotype and environment on the microbiome of the coral Acropora tenuis. PeerJ 7:e6377
Hansson L, Agis M, Maier C, Weinbauer MG (2009) Community composition of bacteria associated with cold-water coral Madrepora oculata: within and between colony variability. Mar Ecol Prog Ser 397:89–102
Hernandez-Agreda A, Gates RD, Ainsworth TD (2016) Defining the core microbiome in corals’ microbial soup. Trends Microbiol 25:125–140
Hernandez-Agreda A, Leggat W, Bongaerts P, Herrera C, Ainsworth T (2018) Rethinking the coral microbiome: simplicity exists within a diverse microbial biosphere. mBio 9:e00812–e00818
Hester ER, Barott KL, Nulton J, Vermeij MJ, Rohwer FL (2016) Stable and sporadic symbiotic communities of coral and algal holobionts. ISME J 10:1157–1169
Horn M, Harzenetter MD, Linner T, Schmid EN, Müller K-D, Michel R, Wagner M (2001) Members of the Cytophaga–Flavobacterium–Bacteroides phylum as intracellular bacteria of acanthamoebae: proposal of ‘Candidatus Amoebophilus asiaticus’. Environ Microbiol 3:440–449
Hothorn T, Bretz F, Westfall P (2008) Simultaneous inference in general parametric models. Biom J 50:346–363
Hughes JB, Hellmann JJ (2005) The application of rarefaction techniques to molecular inventories of microbial diversity. Methods Enzymol 397:292–308
Kemp DW, Rivers AR, Kemp KM, Lipp EK, Porter JW, Wares JP (2015) Spatial homogeneity of bacterial communities associated with the surface mucus layer of the reef-building coral Acropora palmata. PLoS ONE 10:e0143790
Lande R (1996) Statistics and partitioning of species diversity, and similarity among multiple communities. Oikos 76:5–13
Lee MD, Walworth NG, Sylvan JB, Edwards KJ, Orcutt BN (2015) Microbial communities on seafloor basalts at Dorado outcrop reflect level of alteration and highlight global lithic clades. Front Microbiol 6:1470
Lee OO, Yang J, Bougouffa S, Wang Y, Batang Z, Tian R, Al-Suwailem A, Qian PY (2012) Spatial and species variations in bacterial communities associated with corals from the Red Sea as revealed by pyrosequencing. Appl Environ Microbiol 78:7173–7184
Legendre P, Legendre L (1998) Numerical ecology (developments in environmental modelling). Elsevier, Amsterdam
Littman RA, Willis BL, Pfeffer C, Bourne DG (2009) Diversities of coral-associated bacteria differ with location, but not species, for three acroporid corals on the Great Barrier Reef. FEMS Microbiol Ecol 68:152–163
Marcelino VR, van Oppen MJ, Verbruggen H (2018) Highly structured prokaryote communities exist within the skeleton of coral colonies. ISME J 12:300–303
McMurdie PJ, Holmes S (2013) phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 8:e61217
Neave MJ, Apprill A, Ferrier-Pages C, Voolstra CR (2016) Diversity and function of prevalent symbiotic marine bacteria in the genus Endozoicomonas. Appl Microbiol Biotechnol 100:8315–8324
Neave MJ, Michell CT, Apprill A, Voolstra CR (2017) Endozoicomonas genomes reveal functional adaptation and plasticity in bacterial strains symbiotically associated with diverse marine hosts. Scientific Rep 7:40579
Oksanen J, Blanchet FG, Friendly M, Kindt R, Legendre P, McGlinn D, Minchin PR, O’Hara RB, Simpson CL, Solymos P, Henry M, Stevens H, Szoecs E, Wagner H (2016) Vegan: community ecology package. R package version 2.4-1
Pantos O, Bongaerts P, Dennis PG, Tyson GW, Hoegh-Guldberg O (2015) Habitat-specific environmental conditions primarily control the microbiomes of the coral Seriatopora hystrix. ISME J 9:1916–1927
Pollock FJ, McMinds R, Smith S, Bourne DG, Willis BL, Medina M, Vega Thurber R, Zaneveld JR (2018) Coral-associated bacteria demonstrate phylosymbiosis and cophylogeny. Nat Commun 9:1–13
Pratte ZA, Longo GO, Burns AS, Hay ME, Stewart FJ (2018) Contact with turf algae alters the coral microbiome: contact versus systemic impacts. Coral Reefs 37:1–13
Pratte ZA, Richardson LL, Mills DK (2015) Microbiota shifts in the surface mucopolysaccharide layer of corals transferred from natural to aquaria settings. J Invertebr Pathol 125:42–44
Putnam HM, Barott KL, Ainsworth TD, Gates RD (2017) The vulnerability and resilience of reef-building corals. Curr Biol CB 27:R528–R540
QIIME 2 Development Team (2017a) q2-demux. https://github.com/qiime2/q2-demux
QIIME 2 Development Team (2017b) q2-feature-classifier. https://github.com/qiime2/q2-feature-classifier
Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glockner FO (2013) The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41:D590–D596
R Core Team (2018) R: a language and environment for statistical computing. http://www.R-project.org
Raina JB, Tapiolas D, Willis BL, Bourne DG (2009) Coral-associated bacteria and their role in the biogeochemical cycling of sulfur. Appl Environ Microbiol 75:3492–3501
Rohwer F, Seguritan V, Azam F, Knowlton N (2002) Diversity and distribution of coral-associated bacteria. Mar Ecol Prog Ser 243:1–10
Singer E, Wagner M, Woyke T (2017) Capturing the genetic makeup of the active microbiome in situ. ISME J 11:1949–1963
Sweet MJ, Croquer A, Bythell JC (2011) Bacterial assemblages differ between compartments within the coral holobiont. Coral Reefs 30:39–52
Thurber RV, Willner-Hall D, Rodriguez-Mueller B, Desnues C, Edwards RA, Angly F, Dinsdale E, Kelly L, Rohwer F (2009) Metagenomic analysis of stressed coral holobionts. Environ Microbiol 11:2148–2163
Tremblay P, Weinbauer MG, Rottier C, Guérardel Y, Nozais C, Ferrier-Pagès C (2011) Mucus composition and bacterial communities associated with the tissue and skeleton of three scleractinian corals maintained under culture conditions. J Mar Biol Assoc UK 91:649–657
Veron JEN (2000) Corals of the world. Australian Institute of Marine Science, Townsville
Vouga M, Baud D, Greub G (2017) Simkania negevensis, an insight into the biology and clinical importance of a novel member of the Chlamydiales order. Crit Rev Microbiol 43:62–80
Weiss S, Xu ZZ, Peddada S, Amir A, Bittinger K, Gonzalez A, Lozupone C, Zaneveld JR, Vazquez-Baeza Y, Birmingham A, Hyde ER, Knight R (2017) Normalization and microbial differential abundance strategies depend upon data characteristics. Microbiome 5:27
Wickham H (2009) ggplot2: elegant graphics for data analysis. Springer, New York
Wickham H (2017) tidyverse: easily install and load the “Tidyverse”. R package version 1.2.1
Williams AD, Brown BE, Putchim L, Sweet MJ (2015) Age-related shifts in bacterial diversity in a reef coral. PLoS ONE 10:e0144902
Acknowledgements
We thank Grant Milton and Craig Humphrey for coral collection in the field and the National Sea Simulator team at AIMS for maintaining the corals in aquaria. This work was funded by Australian Research Council Laureate Fellowship FL180100036 to MJHvO.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Additional information
Topic Editor Carly Kenkel
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Damjanovic, K., Blackall, L.L., Peplow, L.M. et al. Assessment of bacterial community composition within and among Acropora loripes colonies in the wild and in captivity. Coral Reefs 39, 1245–1255 (2020). https://doi.org/10.1007/s00338-020-01958-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00338-020-01958-y