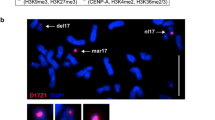
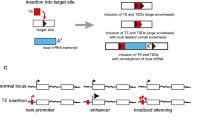
Abstract
The centromere is a chromosomal structure that is essential for the accurate segregation of replicated eukaryotic chromosomes to daughter cells. In most centromeres, the underlying DNA is principally made up of repetitive DNA elements, such as tandemly repeated satellite DNA and retrotransposable elements. Paradoxically, for such an essential genomic region, the DNA is rapidly evolving both within and between species. In this review, we show that the centromere locus is a resilient structure that can undergo evolutionary cycles of birth, growth, maturity, death and resurrection. The birth phase is highlighted by examples in humans and other organisms where centromere DNA deletions or chromosome rearrangements can trigger the epigenetic assembly of neocentromeres onto genomic sites without typical features of centromere DNA. In addition, functional centromeres can be generated in the laboratory using various methodologies. Recent mapping of the foundation centromere mark, the histone H3 variant CENP-A, onto near-complete genomes has uncovered examples of new centromeres which have not accumulated centromere repeat DNA. During the growth period of the centromere, repeat DNA begins to appear at some, but not all, loci. The maturity stage is characterised by centromere repeat accumulation, expansions and contractions and the rapid evolution of the centromere DNA between chromosomes of the same species and between species. This stage provides inherent centromere stability, facilitated by repression of gene activity and meiotic recombination at and around the centromeres. Death to a centromere can result from genomic instability precipitating rearrangements, deletions, accumulation of mutations and the loss of essential centromere binding proteins. Surprisingly, ancestral centromeres can undergo resurrection either in the field or in the laboratory, via as yet poorly understood mechanisms. The underlying principle for the preservation of a centromeric evolutionary life cycle is to provide resilience and perpetuity for the all-important structure and function of the centromere.

Similar content being viewed by others
References
Allshire RC, Karpen GH (2008) Epigenetic regulation of centromeric chromatin: old dogs, new tricks? Nat Rev Genet 9(12):923–937
Amato A, Schillaci T, Lentini L, Di Leonardo A (2009) CENPA overexpression promotes genome instability in pRb-depleted human cells. Mol Cancer 8:119
Amor DJ, Kalitsis P, Sumer H, Choo KHA (2004) Building the centromere: from foundation proteins to 3D organization. Trends Cell Biol 14(7):359–368
Au WC, Crisp MJ, DeLuca SZ, Rando OJ, Basrai MA (2008) Altered dosage and mislocalization of histone H3 and Cse4p lead to chromosome loss in Saccharomyces cerevisiae. Genetics 179(1):263–275
Baker RE, Rogers K (2006) Phylogenetic analysis of fungal centromere H3 proteins. Genetics 174(3):1481–1492
Baldini A, Ried T, Shridhar V, Ogura K, D'Aiuto L, Rocchi M, Ward DC (1993) An alphoid DNA sequence conserved in all human and great ape chromosomes: evidence for ancient centromeric sequences at human chromosomal regions 2q21 and 9q13. Hum Genet 90(6):577–583
Barnhart MC, Kuich PH, Stellfox ME, Ward JA, Bassett EA, Black BE, Foltz DR (2011) HJURP is a CENP-A chromatin assembly factor sufficient to form a functional de novo kinetochore. J Cell Biol 194(2):229–243
Basu J, Stromberg G, Compitello G, Willard HF, Van Bokkelen G (2005) Rapid creation of BAC-based human artificial chromosome vectors by transposition with synthetic alpha-satellite arrays. Nucleic Acids Res 33(2):587–596
Baum M, Sanyal K, Mishra PK, Thaler N, Carbon J (2006) Formation of functional centromeric chromatin is specified epigenetically in Candida albicans. Proc Natl Acad Sci U S A 103(40):14877–14882
Black BE, Cleveland DW (2011) Epigenetic centromere propagation and the nature of CENP-a nucleosomes. Cell 144(4):471–479
Blower MD, Sullivan BA, Karpen GH (2002) Conserved organization of centromeric chromatin in flies and humans. Dev Cell 2(3):319–330
Brinkley BR, Valdivia MM, Tousson A, Brenner SL (1984) Compound kinetochores of the Indian muntjac. Evolution by linear fusion of unit kinetochores. Chromosoma 91(1):1–11
Capozzi O, Purgato S, D'Addabbo P, Archidiacono N, Battaglia P, Baroncini A, Capucci A, Stanyon R, Della Valle G, Rocchi M (2009) Evolutionary descent of a human chromosome 6 neocentromere: a jump back to 17 million years ago. Genome Res 19(5):778–784
Carbone L, Nergadze SG, Magnani E, Misceo D, Francesca Cardone M, Roberto R, Bertoni L, Attolini C, Francesca Piras M, de Jong P, Raudsepp T, Chowdhary BP, Guerin G, Archidiacono N, Rocchi M, Giulotto E (2006) Evolutionary movement of centromeres in horse, donkey, and zebra. Genomics 87(6):777–782
Cellamare A, Catacchio CR, Alkan C, Giannuzzi G, Antonacci F, Cardone MF, Della Valle G, Malig M, Rocchi M, Eichler EE, Ventura M (2009) New insights into centromere organization and evolution from the white-cheeked gibbon and marmoset. Mol Biol Evol 26(8):1889–1900
Choo KHA (1997) The centromere. Oxford University Press, Oxford
Coghlan A, Eichler EE, Oliver SG, Paterson AH, Stein L (2005) Chromosome evolution in eukaryotes: a multi-kingdom perspective. Trends Genet 21(12):673–682
Collins KA, Furuyama S, Biggins S (2004) Proteolysis contributes to the exclusive centromere localization of the yeast Cse4/CENP-A histone H3 variant. Curr Biol 14(21):1968–1972
Cooper JL, Henikoff S (2004) Adaptive evolution of the histone fold domain in centromeric histones. Mol Biol Evol 21(9):1712–1718
Dani GM, Zakian VA (1983) Mitotic and meiotic stability of linear plasmids in yeast. Proc Natl Acad Sci U S A 80(11):3406–3410
Dover G (1982) Molecular drive: a cohesive mode of species evolution. Nature 299(5879):111–117
Dutrillaux B, Rethore MO, Lejeune J (1975) Comparison of the karyotype of the orangutan (Pongo pygmaeus) to those of man, chimpanzee, and gorilla. Ann Genet 18(3):153–161
Earnshaw WC, Migeon BR (1985) Three related centromere proteins are absent from the inactive centromere of a stable isodicentric chromosome. Chromosoma 92(4):290–296
Elde NC, Roach KC, Yao MC, Malik HS (2011) Absence of positive selection on centromeric histones in Tetrahymena suggests unsuppressed centromere: drive in lineages lacking male meiosis. J Mol Evol 72(5–6):510–520
Fitzgerald-Hayes M, Clarke L, Carbon J (1982) Nucleotide sequence comparisons and functional analysis of yeast centromere DNAs. Cell 29(1):235–244
Gascoigne KE, Takeuchi K, Suzuki A, Hori T, Fukagawa T, Cheeseman IM (2011) Induced ectopic kinetochore assembly bypasses the requirement for CENP-A nucleosomes. Cell 145(3):410–422
Guerra M, Cabral G, Cuacos M, Gonzalez-Garcia M, Gonzalez-Sanchez M, Vega J, Puertas MJ (2010) Neocentrics and holokinetics (holocentrics): chromosomes out of the centromeric rules. Cytogenet Genome Res 129(1–3):82–96
Haaf T, Ward DC (1994) Structural analysis of alpha-satellite DNA and centromere proteins using extended chromatin and chromosomes. Hum Mol Genet 3(5):697–709
Haaf T, Warburton PE, Willard HF (1992) Integration of human alpha-satellite DNA into simian chromosomes: centromere protein binding and disruption of normal chromosome segregation. Cell 70(4):681–696
Hagstrom KA, Holmes VF, Cozzarelli NR, Meyer BJ (2002) C. elegans condensin promotes mitotic chromosome architecture, centromere organization, and sister chromatid segregation during mitosis and meiosis. Genes Dev 16(6):729–742
Hahnenberger KM, Baum MP, Polizzi CM, Carbon J, Clarke L (1989) Construction of functional artificial minichromosomes in the fission yeast Schizosaccharomyces pombe. Proc Natl Acad Sci U S A 86(2):577–581
Han F, Lamb JC, Birchler JA (2006) High frequency of centromere inactivation resulting in stable dicentric chromosomes of maize. Proc Natl Acad Sci U S A 103(9):3238–3243
Han Y, Zhang Z, Liu C, Liu J, Huang S, Jiang J, Jin W (2009) Centromere repositioning in cucurbit species: implication of the genomic impact from centromere activation and inactivation. Proc Natl Acad Sci U S A 106(35):14937–14941
Harrington JJ, Van Bokkelen G, Mays RW, Gustashaw K, Willard HF (1997) Formation of de novo centromeres and construction of first-generation human artificial microchromosomes. Nat Genet 15(4):345–355
Hassold T, Hunt P (2001) To err (meiotically) is human: the genesis of human aneuploidy. Nat Rev Genet 2(4):280–291
Henikoff S, Ahmad K, Malik HS (2001) The centromere paradox: stable inheritance with rapidly evolving DNA. Science 293(5532):1098–1102
Heun P, Erhardt S, Blower MD, Weiss S, Skora AD, Karpen GH (2006) Mislocalization of the Drosophila centromere-specific histone CID promotes formation of functional ectopic kinetochores. Dev Cell 10(3):303–315
Heus JJ, Zonneveld BJ, Steensma HY, Van den Berg JA (1990) Centromeric DNA of Kluyveromyces lactis. Curr Genet 18(6):517–522
Heus JJ, Zonneveld BJ, de Steensma HY, van den Berg JA (1993) The consensus sequence of Kluyveromyces lactis centromeres shows homology to functional centromeric DNA from Saccharomyces cerevisiae. Mol Gen Genet 236(2–3):355–362
Higgins AW, Gustashaw KM, Willard HF (2005) Engineered human dicentric chromosomes show centromere plasticity. Chromosome Res 13(8):745–762
Hori T, Amano M, Suzuki A, Backer CB, Welburn JP, Dong Y, McEwen BF, Shang WH, Suzuki E, Okawa K, Cheeseman IM, Fukagawa T (2008) CCAN makes multiple contacts with centromeric DNA to provide distinct pathways to the outer kinetochore. Cell 135(6):1039–1052
Hudson DF, Fowler KJ, Earle E, Saffery R, Kalitsis P, Trowell H, Hill J, Wreford NG, de Kretser DM, Cancilla MR, Howman E, Hii L, Cutts SM, Irvine DV, Choo KHA (1998) Centromere protein B null mice are mitotically and meiotically normal but have lower body and testis weights. J Cell Biol 141(2):309–319
Ikeno M, Grimes B, Okazaki T, Nakano M, Saitoh K, Hoshino H, McGill NI, Cooke H, Masumoto H (1998) Construction of YAC-based mammalian artificial chromosomes. Nat Biotechnol 16(5):431–439
Irvine DV, Amor DJ, Perry J, Sirvent N, Pedeutour F, Choo KHA, Saffery R (2004) Chromosome size and origin as determinants of the level of CENP-A incorporation into human centromeres. Chromosome Res 12(8):805–815
Ishii K, Ogiyama Y, Chikashige Y, Soejima S, Masuda F, Kakuma T, Hiraoka Y, Takahashi K (2008) Heterochromatin integrity affects chromosome reorganization after centromere dysfunction. Science 321(5892):1088–1091
Kapoor M, de Oca M, Luna R, Liu G, Lozano G, Cummings C, Mancini M, Ouspenski I, Brinkley BR, May GS (1998) The cenpB gene is not essential in mice. Chromosoma 107(8):570–576
Ketel C, Wang HS, McClellan M, Bouchonville K, Selmecki A, Lahav T, Gerami-Nejad M, Berman J (2009) Neocentromeres form efficiently at multiple possible loci in Candida albicans. PLoS Genet 5(3):e1000400
Kipling D, Warburton PE (1997) Centromeres, CENP-B and Tigger too. Trends Genet 13(4):141–145
Kitada K, Yamaguchi E, Arisawa M (1996) Isolation of a Candida glabrata centromere and its use in construction of plasmid vectors. Gene 175(1–2):105–108
Koshland D, Rutledge L, Fitzgerald-Hayes M, Hartwell LH (1987) A genetic analysis of dicentric minichromosomes in Saccharomyces cerevisiae. Cell 48(5):801–812
Lam AL, Boivin CD, Bonney CF, Rudd MK, Sullivan BA (2006) Human centromeric chromatin is a dynamic chromosomal domain that can spread over noncentromeric DNA. Proc Natl Acad Sci U S A 103(11):4186–4191
Larin Z, Fricker MD, Tyler-Smith C (1994) De novo formation of several features of a centromere following introduction of a Y alphoid YAC into mammalian cells. Hum Mol Genet 3(5):689–695
Lee HR, Zhang W, Langdon T, Jin W, Yan H, Cheng Z, Jiang J (2005) Chromatin immunoprecipitation cloning reveals rapid evolutionary patterns of centromeric DNA in Oryza species. Proc Natl Acad Sci U S A 102(33):11793–11798
Lo AWI, Craig JM, Saffery R, Kalitsis P, Irvine DV, Earle E, Magliano DJ, Choo KHA (2001) A 330 kb CENP-A binding domain and altered replication timing at a human neocentromere. EMBO J 20(8):2087–2096
Locke DP, Hillier LW, Warren WC, Worley KC, Nazareth LV, Muzny DM, Yang SP, Wang Z, Chinwalla AT, Minx P, Mitreva M, Cook L, Delehaunty KD, Fronick C, Schmidt H, Fulton LA, Fulton RS, Nelson JO, Magrini V, Pohl C, Graves TA, Markovic C, Cree A, Dinh HH, Hume J, Kovar CL, Fowler GR, Lunter G, Meader S, Heger A, Ponting CP, Marques-Bonet T, Alkan C, Chen L, Cheng Z, Kidd JM, Eichler EE, White S, Searle S, Vilella AJ, Chen Y, Flicek P, Ma J, Raney B, Suh B, Burhans R, Herrero J, Haussler D, Faria R, Fernando O, Darre F, Farre D, Gazave E, Oliva M, Navarro A, Roberto R, Capozzi O, Archidiacono N, Della Valle G, Purgato S, Rocchi M, Konkel MK, Walker JA, Ullmer B, Batzer MA, Smit AF, Hubley R, Casola C, Schrider DR, Hahn MW, Quesada V, Puente XS, Ordonez GR, Lopez-Otin C, Vinar T, Brejova B, Ratan A, Harris RS, Miller W, Kosiol C, Lawson HA, Taliwal V, Martins AL, Siepel A, Roychoudhury A, Ma X, Degenhardt J, Bustamante CD, Gutenkunst RN, Mailund T, Dutheil JY, Hobolth A, Schierup MH, Ryder OA, Yoshinaga Y, de Jong PJ, Weinstock GM, Rogers J, Mardis ER, Gibbs RA, Wilson RK (2011) Comparative and demographic analysis of orang-utan genomes. Nature 469(7331):529–533
Lukaszewski AJ (1995) Chromatid and chromosome type breakage-fusion-bridge cycles in wheat (Triticum aestivum L.). Genetics 140(3):1069–1085
Ma J, Wing RA, Bennetzen JL, Jackson SA (2007) Plant centromere organization: a dynamic structure with conserved functions. Trends Genet 23(3):134–139
Malik HS (2009) The centromere-drive hypothesis: a simple basis for centromere complexity. Prog Mol Subcell Biol 48:33–52
Malik HS, Henikoff S (2001) Adaptive evolution of Cid, a centromere-specific histone in Drosophila. Genetics 157(3):1293–1298
Malik HS, Henikoff S (2009) Major evolutionary transitions in centromere complexity. Cell 138(6):1067–1082
Marshall OJ, Choo KH (2009) Neocentromeres come of age. PLoS Genet 5(3):e1000370
Marshall OJ, Chueh AC, Wong LH, Choo KHA (2008a) Neocentromeres: new insights into centromere structure, disease development, and karyotype evolution. Am J Hum Genet 82(2):261–282
Marshall OJ, Marshall AT, Choo KHA (2008b) Three-dimensional localization of CENP-A suggests a complex higher order structure of centromeric chromatin. J Cell Biol 183(7):1193–1202
Masumoto H, Masukata H, Muro Y, Nozaki N, Okazaki T (1989) A human centromere antigen (CENP-B) interacts with a short specific sequence in alphoid DNA, a human centromeric satellite. J Cell Biol 109(5):1963–1973
McClintock B (1938) The production of homozygous deficient tissues with mutant characteristics by means of the aberrant mitotic behaviour of ring-shaped chromosomes. Genetics 23:315–376
Mendiburo MJ, Padeken J, Fulop S, Schepers A, Heun P (2011) Drosophila CENH3 is sufficient for centromere formation. Science 334(6056):686–690
Meraldi P, McAinsh AD, Rheinbay E, Sorger PK (2006) Phylogenetic and structural analysis of centromeric DNA and kinetochore proteins. Genome Biol 7(3):R23
Miller DA, Sharma V, Mitchell AR (1988) A human-derived probe, p82H, hybridizes to the centromeres of gorilla, chimpanzee, and orangutan. Chromosoma 96(4):270–274
Misceo D, Capozzi O, Roberto R, Dell'oglio MP, Rocchi M, Stanyon R, Archidiacono N (2008) Tracking the complex flow of chromosome rearrangements from the Hominoidea Ancestor to extant Hylobates and Nomascus Gibbons by high-resolution synteny mapping. Genome Res 18(9):1530–1537
Montefalcone G, Tempesta S, Rocchi M, Archidiacono N (1999) Centromere repositioning. Genome Res 9(12):1184–1188
Moreno-Moreno O, Torras-Llort M, Azorin F (2006) Proteolysis restricts localization of CID, the centromere-specific histone H3 variant of Drosophila, to centromeres. Nucleic Acids Res 34(21):6247–6255
Nagaki K, Talbert PB, Zhong CX, Dawe RK, Henikoff S, Jiang J (2003) Chromatin immunoprecipitation reveals that the 180-bp satellite repeat is the key functional DNA element of Arabidopsis thaliana centromeres. Genetics 163(3):1221–1225
Nagaki K, Cheng Z, Ouyang S, Talbert PB, Kim M, Jones KM, Henikoff S, Buell CR, Jiang J (2004) Sequencing of a rice centromere uncovers active genes. Nat Genet 36(2):138–145
Nagaki K, Walling J, Hirsch C, Jiang J, Murata M (2009) Structure and evolution of plant centromeres. Prog Mol Subcell Biol 48:153–179
Nasuda S, Hudakova S, Schubert I, Houben A, Endo TR (2005) Stable barley chromosomes without centromeric repeats. Proc Natl Acad Sci U S A 102(28):9842–9847
Ohkuma M, Kobayashi K, Kawai S, Hwang CW, Ohta A, Takagi M (1995) Identification of a centromeric activity in the autonomously replicating TRA region allows improvement of the host-vector system for Candida maltosa. Mol Gen Genet 249(4):447–455
Ohzeki J, Nakano M, Okada T, Masumoto H (2002) CENP-B box is required for de novo centromere chromatin assembly on human alphoid DNA. J Cell Biol 159(5):765–775
Okada T, Ohzeki J, Nakano M, Yoda K, Brinkley WR, Larionov V, Masumoto H (2007) CENP-B controls centromere formation depending on the chromatin context. Cell 131(7):1287–1300
Olszak AM, van Essen D, Pereira AJ, Diehl S, Manke T, Maiato H, Saccani S, Heun P (2011) Heterochromatin boundaries are hotspots for de novo kinetochore formation. Nat Cell Biol 13(7):799–808
Page SL, Earnshaw WC, Choo KHA, Shaffer LG (1995) Further evidence that CENP-C is a necessary component of active centromeres: studies of a dic(X; 15) with simultaneous immunofluorescence and FISH. Hum Mol Genet 4(2):289–294
Perez-Castro AV, Shamanski FL, Meneses JJ, Lovato TL, Vogel KG, Moyzis RK, Pedersen R (1998) Centromeric protein B null mice are viable with no apparent abnormalities. Dev Biol 201(2):135–143
Perpelescu M, Fukagawa T (2011) The ABCs of CENPs. Chromosoma 120:425–446
Pertile MD, Graham AN, Choo KHA, Kalitsis P (2009) Rapid evolution of mouse Y centromere repeat DNA belies recent sequence stability. Genome Res 19(12):2202–2213
Platero JS, Ahmad K, Henikoff S (1999) A distal heterochromatic block displays centromeric activity when detached from a natural centromere. Mol Cell 4(6):995–1004
Plohl M, Luchetti A, Mestrovic N, Mantovani B (2008) Satellite DNAs between selfishness and functionality: structure, genomics and evolution of tandem repeats in centromeric (hetero)chromatin. Gene 409(1–2):72–82
Ranjitkar P, Press MO, Yi X, Baker R, MacCoss MJ, Biggins S (2010) An E3 ubiquitin ligase prevents ectopic localization of the centromeric histone H3 variant via the centromere targeting domain. Mol Cell 40(3):455–464
Ribeiro SA, Gatlin JC, Dong Y, Joglekar A, Cameron L, Hudson DF, Farr CJ, McEwen BF, Salmon ED, Earnshaw WC, Vagnarelli P (2009) Condensin regulates the stiffness of vertebrate centromeres. Mol Biol Cell 20(9):2371–2380
Rocchi M, Archidiacono N, Schempp W, Capozzi O, Stanyon R (2012) Centromere repositioning in mammals. Hered (Edinb) 108(1):59–67
Rudd MK, Willard HF (2004) Analysis of the centromeric regions of the human genome assembly. Trends Genet 20(11):529–533
Rudd MK, Mays RW, Schwartz S, Willard HF (2003a) Human artificial chromosomes with alpha satellite-based de novo centromeres show increased frequency of nondisjunction and anaphase lag. Mol Cell Biol 23(21):7689–7697
Rudd MK, Schueler MG, Willard HF (2003b) Sequence organization and functional annotation of human centromeres. Cold Spring Harb Symp Quant Biol 68:141–149
Santaguida S, Musacchio A (2009) The life and miracles of kinetochores. EMBO J 28:2511–2531
Schueler MG, Swanson W, Thomas PJ, Green ED (2010) Adaptive evolution of foundation kinetochore proteins in primates. Mol Biol Evol 27(7):1585–1597
Scott KC, Merrett SL, Willard HF (2006) A heterochromatin barrier partitions the fission yeast centromere into discrete chromatin domains. Curr Biol 16(2):119–129
Seuanez H, Fletcher J, Evans HJ, Martin DE (1976) A chromosome rearrangement in orangutan studied with Q-, C-, and G-banding techniques. Cytogenet Cell Genet 17(1):26–34
Shang WH, Hori T, Toyoda A, Kato J, Popendorf K, Sakakibara Y, Fujiyama A, Fukagawa T (2010) Chickens possess centromeres with both extended tandem repeats and short non-tandem-repetitive sequences. Genome Res 20(9):1219–1228
She X, Horvath JE, Jiang Z, Liu G, Furey TS, Christ L, Clark R, Graves T, Gulden CL, Alkan C, Bailey JA, Sahinalp C, Rocchi M, Haussler D, Wilson RK, Miller W, Schwartz S, Eichler EE (2004) The structure and evolution of centromeric transition regions within the human genome. Nature 430(7002):857–864
Shi J, Wolf SE, Burke JM, Presting GG, Ross-Ibarra J, Dawe RK (2010) Widespread gene conversion in centromere cores. PLoS Biol 8(3):e1000327
Smit AFA, Riggs AD (1996) Tiggers and DNA transposon fossils in the human genome. Proc Natl Acad Sci U S A 93(4):1443–1448
Smith GP (1976) Evolution of repeated DNA sequences by unequal crossover. Science 191(4227):528–535
Smith KM, Phatale PA, Sullivan CM, Pomraning KR, Freitag M (2011) Heterochromatin is required for normal distribution of Neurospora crassa CenH3. Mol Cell Biol 31(12):2528–2542
Southern EM (1975) Long range periodicities in mouse satellite DNA. J Mol Biol 94(1):51–69
Steiner NC, Clarke L (1994) A novel epigenetic effect can alter centromere function in fission yeast. Cell 79(5):865–874
Stoyan T, Carbon J (2004) Inner kinetochore of the pathogenic yeast Candida glabrata. Eukaryot Cell 3(5):1154–1163
Sullivan BA, Schwartz S (1995) Identification of centromeric antigens in dicentric Robertsonian translocations: CENP-C and CENP-E are necessary components of functional centromeres. Hum Mol Genet 4(12):2189–2197
Sun X, Wahlstrom J, Karpen G (1997) Molecular structure of a functional Drosophila centromere. Cell 91(7):1007–1019
Talbert PB, Masuelli R, Tyagi AP, Comai L, Henikoff S (2002) Centromeric localization and adaptive evolution of an Arabidopsis histone H3 variant. Plant Cell 14(5):1053–1066
Talbert PB, Bryson TD, Henikoff S (2004) Adaptive evolution of centromere proteins in plants and animals. J Biol 3(4):18
Thompson SL, Bakhoum SF, Compton DA (2010) Mechanisms of chromosomal instability. Curr Biol 20(6):R285–R295
Tomonaga T, Matsushita K, Yamaguchi S, Oohashi T, Shimada H, Ochiai T, Yoda K, Nomura F (2003) Overexpression and mistargeting of centromere protein-A in human primary colorectal cancer. Cancer Res 63(13):3511–3516
Vafa O, Shelby RD, Sullivan KF (1999) CENP-A associated complex satellite DNA in the kinetochore of the Indian muntjac. Chromosoma 108(6):367–374
Van Hooser AA, Ouspenski II, Gregson HC, Starr DA, Yen TJ, Goldberg ML, Yokomori K, Earnshaw WC, Sullivan KF, Brinkley BR (2001) Specification of kinetochore-forming chromatin by the histone H3 variant CENP-A. J Cell Sci 114(Pt 19):3529–3542
Ventura M, Mudge JM, Palumbo V, Burn S, Blennow E, Pierluigi M, Giorda R, Zuffardi O, Archidiacono N, Jackson MS, Rocchi M (2003) Neocentromeres in 15q24-26 map to duplicons which flanked an ancestral centromere in 15q25. Genome Res 13(9):2059–2068
Wade CM, Giulotto E, Sigurdsson S, Zoli M, Gnerre S, Imsland F, Lear TL, Adelson DL, Bailey E, Bellone RR, Blocker H, Distl O, Edgar RC, Garber M, Leeb T, Mauceli E, MacLeod JN, Penedo MC, Raison JM, Sharpe T, Vogel J, Andersson L, Antczak DF, Biagi T, Binns MM, Chowdhary BP, Coleman SJ, Della Valle G, Fryc S, Guerin G, Hasegawa T, Hill EW, Jurka J, Kiialainen A, Lindgren G, Liu J, Magnani E, Mickelson JR, Murray J, Nergadze SG, Onofrio R, Pedroni S, Piras MF, Raudsepp T, Rocchi M, Roed KH, Ryder OA, Searle S, Skow L, Swinburne JE, Syvanen AC, Tozaki T, Valberg SJ, Vaudin M, White JR, Zody MC, Lander ES, Lindblad-Toh K (2009) Genome sequence, comparative analysis, and population genetics of the domestic horse. Science 326(5954):865–867
Wang H, Xu Z, Gao L, Hao B (2009) A fungal phylogeny based on 82 complete genomes using the composition vector method. BMC Evol Biol 9:195
Warburton PE (2004) Chromosomal dynamics of human neocentromere formation. Chromosome Res 12(6):617–626
Weaver BA, Cleveland DW (2007) Aneuploidy: instigator and inhibitor of tumorigenesis. Cancer Res 67(21):10103–10105
Williams BC, Murphy TD, Goldberg ML, Karpen GH (1998) Neocentromere activity of structurally acentric mini-chromosomes in Drosophila. Nat Genet 18(1):30–37
Yan H, Talbert PB, Lee HR, Jett J, Henikoff S, Chen F, Jiang J (2008) Intergenic locations of rice centromeric chromatin. PLoS Biol 6(11):e286
Yunis JJ, Prakash O (1982) The origin of man: a chromosomal pictorial legacy. Science 215(4539):1525–1530
Zhong CX, Marshall JB, Topp C, Mroczek R, Kato A, Nagaki K, Birchler JA, Jiang J, Dawe RK (2002) Centromeric retroelements and satellites interact with maize kinetochore protein CENH3. Plant Cell 14(11):2825–2836
Zinkowski RP, Meyne J, Brinkley BR (1991) The centromere–kinetochore complex: a repeat subunit model. J Cell Biol 113(5):1091–1110
Acknowledgments
This work was supported by the National Health and Medical Research Council of Australia and the Victorian Government’s Operational Infrastructure Support Programme. PK was supported by an R.D. Wright Fellowship and KHAC by a Senior Principal Research Fellowship of NHMRC.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Erich Nigg
Rights and permissions
About this article
Cite this article
Kalitsis, P., Choo, K.H.A. The evolutionary life cycle of the resilient centromere. Chromosoma 121, 327–340 (2012). https://doi.org/10.1007/s00412-012-0369-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00412-012-0369-6