Abstract
On November 26, 2021, the World Health Organization classified B.1.1.529 as a severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variant of concern (VoC), named omicron. Spike-gene dropouts in conventional SARS-CoV-2 PCR systems have been reported over the last weeks as indirect diagnostic evidence for the identification of omicron. Here, we report the combination of PCRs specific for heavily mutated sites in the spike gene and nanopore-based full-length genome sequencing for the rapid and sensitive identification of the first four COVID-19 patients diagnosed in Germany to be infected with omicron on November 28, 2021. This study will assist the unambiguous laboratory-based diagnosis and global surveillance for this highly contagious VoC with an unprecedented degree of humoral immune escape. Moreover, we propose that specialized diagnostic laboratories should continuously update their assays for variant-specific PCRs in the spike gene of SARS-CoV-2 to readily detect and diagnose emerging variants of interest and VoCs. The combination with established nanopore sequencing procedures allows both the rapid confirmation by whole genome sequencing as well as the sensitive identification of newly emerging variants of this pandemic β-coronavirus in years to come.
Similar content being viewed by others
Introduction
Already in the last weeks of December 2021, omicron has become the dominant variant of concern (VoC) in many countries, including Great Britain, France, Denmark, Greece, Spain, Portugal, and the US [1] with numbers of new infections reaching all-time highs. Earlier VoCs were characterized either by an increased ability for transmission in the population [VoCs alpha (B.1.1.7) and delta (B.1.617.2)] or a partial immune escape with variable effects on neutralization by polyclonal serum antibodies [VoCs beta (B.1.351), gamma (P.1/B.1.1.28) and delta] [2,3,4,5,6,7]. A striking characteristic of the apparently independently evolved omicron VoC is a large number of amino acid substitutions, insertions and deletions in the spike protein, i.e. 32 changes compared to the original Wuhan-hu-1 virus [8], that likely contribute to its extraordinarily rapid spread in the population. Moreover, the number of epitopes in the spike protein relevant for neutralization are an important determinant of the genetic barrier to viral escape from humoral immunity [6, 9]. Consequently, physician-scientists have been alerted since its discovery based on the genetic information alone by omicron’s potential for a pronounced immune escape. Recent laboratory studies and epidemiological reports have now confirmed this exceptional combination of high contagiousity and drastic escape from neutralizing antibodies [10,11,12]. The resistance to neutralization is most likely due to mutations K417N, N440K, G446S, S477N, T478K, E484A, Q493R, G496S, Q498R, N501Y and Y505H, which are located within or close to the epitopes bound by these antibodies [13].
Here, we report the laboratory-based diagnosis of the first four COVID-19 cases with an omicron infection reported in Germany based on a combination of variant-specific PCRs and next-generation-sequencing using an established nanopore platform.
Materials and methods
Patients and collection of respiratory samples for testing for SARS-CoV-2
Respiratory samples (either oropharyngeal gargle or nasopharyngeal swab) were collected in Bio-Speedy vNAT Transfer Tubes (Bioeksen R&D Technologies, Cat. No. BS-NA-513-100) from patients on November 27, 2021, who had self-reported a positive SARS-CoV-2 PCR result two days after returning from Cape Town, South Africa, following a two-week round trip (patient 1 and 2), or patients arriving in Germany at Munich airport on a flight from Cape Town, South Africa, on November 26, 2021 (patient 3 and 4). For additional patient characteristics and sample details see Table 1.
Quantitative viral load determination
Samples were quantified for SARS-CoV-2 RNA using the fully-automated Roche cobas® 6800/8800 system (Roche, Mannheim, Germany). For quantification, standard curves were generated in multiple replicates using a commercially available standard for calibration (Instand e.V.) as described previously [14]. Viral loads of samples were calculated as SARS-CoV-2 nucleocapsid gene copy numbers per 1 ml of oropharyngeal gargle.
SARS-CoV-2 VOC-specific PCRs
Nucleic acids from patients’ respiratory samples were extracted using the QIAsymphony DSP Virus/Pathogen Kit (cat. no. 937036) on a QIAsymphony SP instrument or the EZ1 Virus Mini Kit v2.0 (cat. no. 955134) with the EZ1 Advanced XL (Qiagen, Hilden, Germany). Eluates were used for melting curve analysis performed on a LightCycler480 II (Software version 1.5.1.62) (Roche Diagnostics, Basel, Switzerland) with the Luna® Probe One-Step RT-qPCR Kit (No ROX) (cat. no. E3007E, NEB, Frankfurt am Main, Germany) and the following VirSNiP kits (TIB MOLBIOL, Berlin, Germany): SARS del69/70 + 484 K + 501Y (Cat. No. 53-0799-96) (Fig. 1A, top panel), SARS Spike 417 T 681H (Cat. No. 53-0807-96) (Fig. 1A, middle panel) and SARS Spike S371L S373P (53-0827-96) (Fig. 1a, lower panel). We followed the manufacturer instructions for the VirSNiP assay except adapting the thermocycler program to the Luna® Probe One-Step RT-qPCR mix with the following modification: we shortened the 60 °C step to 15 s and added a step at 72 °C for 15 s in every cycle. Seegene Variants I and II assays (cat. no. RV10286X and RV10305X) were performed on C1000 Thermal Cyclers (Bio-Rad, Feldkirchen, Germany) according to the manufacturer’s instructions.
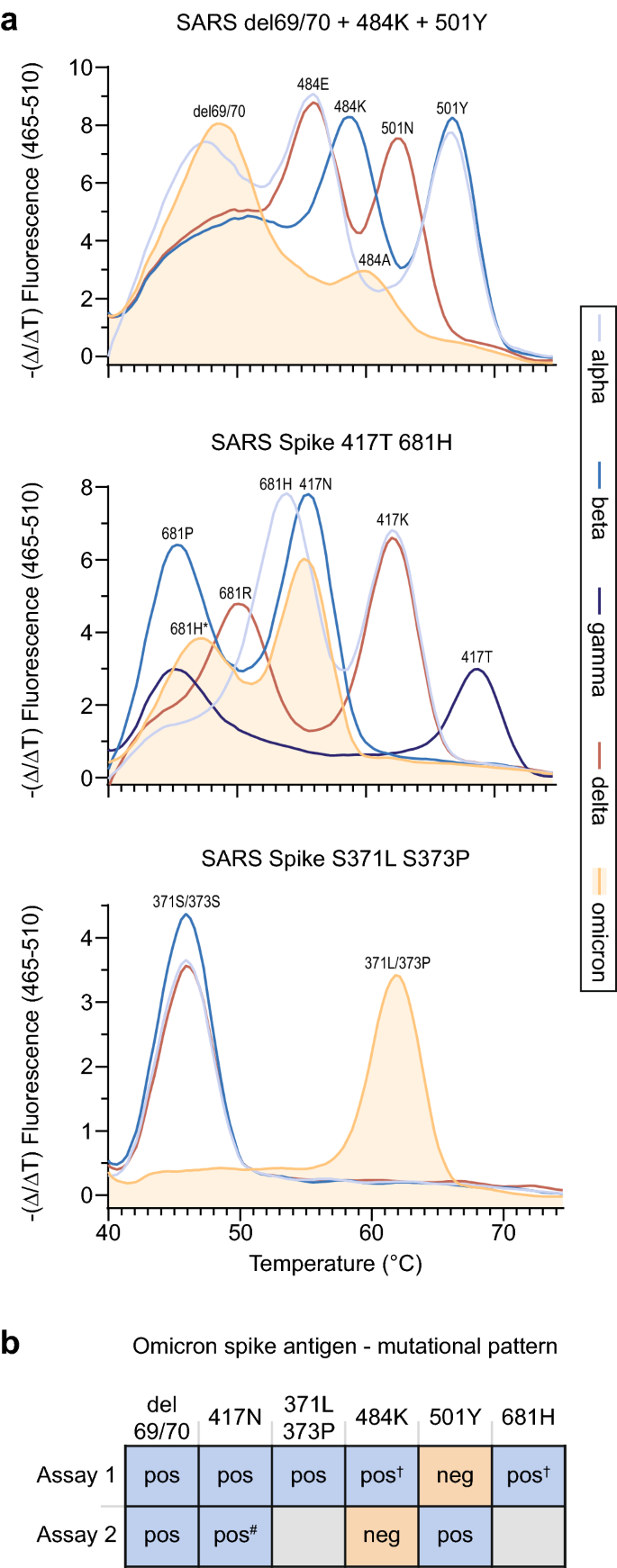
Detection of omicron (B.1.1.529) by SARS-CoV-2 variant-specific PCRs for the spike gene. a PCR probe-based melting curve assays (ref 26) for the identification of SARS-CoV-2 VOCs. Melting curves for spike mutations/deletions at the indicated amino acid positions. Data for all five VOCs are depicted. b Summary of mutational pattern for omicron detected by two different VOC-specific assays (Assay 1: TIB MOLBIOL (ref 26); Assay 2: Seegene Variants I/II (refs 27, 28). † Melting curve peak shifted and dampened. #Delayed Ct value
SARS-CoV-2 whole-genome sequencing
Sequencing libraries were prepared according to the ARTIC network nCoV-2019 sequencing protocol v3 [15, 16]. cDNA was generated from extracted total RNA using LunaScript RT (NEB, Ipswich, USA) and amplified with ARTIC primer set v3 [17]. The v3 primer set was rebalanced to achieve a more uniform amplicon coverage [18] and supplemented with updated versions of primer 64_left (ATGTCGATAGATATCCTGTTAATTCCATTGT) and 72_right (CTAGAATAAACTCCACTTTCCATCCAAC) to avoid lineage-specific dropouts. The library quality was routinely checked in each run using standardised control samples. PCR amplification was as described [15, 16] with 35 cycles and 64 °C annealing temperature. PCR reactions were diluted 1:10 and the two primer pool reactions for each sample were pooled. Diluted amplicons were end-repaired, purified with Ampure XP beads, ligated to barcode adapters (Oxford Nanopore Technologies (ONT), Oxford, UK), purified again with Ampure XP beads, pooled and ligated to AMII sequencing adapter (ONT, Oxford, UK). The purified library was sequenced on a PromethION R9.4.1 flowcell until sufficient coverage for lineage calling was reached, monitored by RAMPART software package [19].
Sequencing reads were basecalled with the Guppy basecaller (v.5.0.17) using the super-accurate DNA model provided by ONT (Oxford, UK). The samples were demultiplexed according to the EXP-NBD196 barcode kit. The sequenced amplicons were assembled using the ARTIC bioinformatics protocol [20]. Following the protocol, the artic tool was used to filter the reads by the quality and by a read length between 380 and 700 nucleotides. Reads were mapped to the Sars-Cov-2 reference genome (NC_045512.2) to call variants. Called variants were integrated into the reference sequence to obtain the consensus sequence of the sequenced sample. The lineage assignment was performed with the pangolin tool (v.3.1.16) [21].
Results and discussion
In four individuals returning from South Africa on November 24 or 26, 2021 to Munich, Germany, SARS-CoV-2 was detected in respiratory swabs by PCR on November 27, 2021 with cycle-threshold (ct) values ranging from 23.2 to 31.2 (Table 1). Spike-gene dropouts (or spike-gene target failure, SGTF) in conventional PCR systems, which can present either as a markedly delayed Ct value for or an entirely absent detection of the SARS-CoV-2 spike gene, have been reported for B.1.1.529 over the last weeks [22,23,24] as indirect diagnostic evidence for its identification. Typically, non-spike gene-targeted sequences, e.g. in the nucleocapsid gene, envelope gene or RNA-dependent RNA polymerase gene [25], performed in these omicron-containing samples in parallel, can be readily amplified.
High-resolution probe-based melting curve assays [26] and multiplex-based PCR assays [27, 28] have allowed simultaneous detection of discriminatory mutation sites for identification and differentiation of VOCs alpha [B.1.1.7], beta [B.1.351], gamma [P.1], and delta [B.1.617.2 and AY]. These tests’ applicability to the identification of omicron [B.1.1.529] had not been reported at the time of this study in late November 2021.
Based on the patients’ recent travel history to South Africa and available sequences for B.1.1.529 from GISAID [29], we employed SARS-CoV-2 variant-specific PCR systems (Assays 1 and 2) from two globally operating commercial vendors (TIB MOLBIOL, Berlin, Germany, and Seegene, Germany, Düsseldorf, Germany) [26,27,28], which had at the time either not been officially launched or only recently been launched, on November 27, 2021: RT-PCR products of samples from patients 1–3 (Table 1) showed a peak for del 69/70 (like alpha; unlike beta, gamma and delta) and a shifted and dampened peak for 484 compared to the other VOCs (Fig. 1a (top panel), Tables 2, and 3). The peak for 501 was lost, likely due to surrounding mutations (G496S, Q498R, Y505H). A peak was detected for 417N (like beta; unlike alpha, gamma and delta), and a shifted and dampened peak for 681H (Fig. 1a (middle panel), Tables 2, and 3) (unlike all other VOCs), the latter likely due to omicron’s N679K mutation. Also, a distinct melting curve for 371L/373P compared to alpha, beta and delta was observed (Fig. 1a (lower panel), Tables 2, and 3) [26]. Results for the sample from patient 4 were indeterminate in both assays (Fig. 1b), likely due to the very low virus load in this respiratory sample (ct value of 31.3 or negative; Table 1). Figure 1b summarizes the results for the VOC-specific assays for omicron (see Table 3 for primary output data for Assay 2 for all VOCs [27, 28]).
Next, RNA extracts of patients’ samples were studied by Nanopore sequencing using the ARTIC protocol version 3 [15, 16], which contains 98 primer pairs designed to cover the entire SARS-CoV-2 genome with overlapping 400-bp amplicons. In less than 12 h, sequencing results and SARS-CoV-2 Pangolin lineage calling were completed (GISAID accession ID in Table 1). Some mutations in the spike gene of the omicron VOC result in mismatches for primers used in amplicon generation. For example, amplicon number 76 of the Artic protocol v3/v4 is affected by spike mutations A23040G, G23048A and A23055G, which results in reduced coverage of the spike region including the amino acid mutations K417N, N440K and G446S. Modified protocols that include additional primers matching these polymorphic sites have been developed and will be implemented in future versions [30]. Of particular note, also the sample of patient 4 with very low/negative virus load as assessed by PCR could now be identified as B.1.1.529 with a genome coverage of 95.04%. This indicates that nanopore sequencing may allow a rapid, highly sensitive determination of the SARS-CoV-2 lineage, including omicron. Compared to established short-read sequencing technologies such as Illumina, the long-read nanopore technology allows more cost-effective processing of smaller batches of samples and offers shorter turnaround times. However, the analytical accuracy of nanopore sequencing is limited for the detection of short indels and variants at low read-count frequencies [31]. For larger batches of samples, where turnaround time is not critical, other sequencing technologies could be more cost-effective.
In conclusion, we report the combination of PCRs specific for heavily mutated sites in the spike gene and nanopore-based full-length genome sequencing for the rapid and sensitive identification of the first four COVID-19 patients diagnosed in Germany to be infected with omicron. These findings will assist unambiguous laboratory-based clinical diagnosis and global surveillance for this highly contagious VOC, which carries spike mutations conferring reduced COVID-19 vaccine efficacy. Moreover, specialized diagnostic laboratories should continuously update their assays for variant-specific PCRs in the spike gene of SARS-CoV-2 to readily detect and diagnose emerging variants of interest and VoCs. The combination with established nanopore sequencing procedures allows both the rapid confirmation by whole genome sequencing as well as the sensitive identification of newly emerging variants of this pandemic β-coronavirus in years to come.
Availability of data and material
Not applicable.
Code availability
Not applicable.
References
Mohapatra RK, Sarangi AK, Kandi V et al (2021) Omicron (B.1.1.529 variant of SARS-CoV-2); an emerging threat: current global scenario. J Med Virol. https://doi.org/10.1002/jmv.27561
Volz E, Mishra S et al (2021) Assessing transmissibility of SARS-CoV-2 lineage B.1.1.7 in England. Nature 593:266–269. https://doi.org/10.1038/s41586-021-03470-x
Tegally H, Wilkinson E, Giovanetti M et al (2021) Detection of a SARS-CoV-2 variant of concern in South Africa. Nature 592:438–443. https://doi.org/10.1038/s41586-021-03402-9
Fujino T, Nomoto H, Kutsuna S et al (2021) Novel SARS-CoV-2 variant in travelers from Brazil to Japan. Emerg Infect Dis. https://doi.org/10.3201/eid2704.210138
Mlcochova P, Kemp SA, Dhar MS et al (2021) SARS-CoV-2 B.1.617.2 delta variant replication and immune evasion. Nature 599:114–119. https://doi.org/10.1038/s41586-021-03944-y
Wang Z, Schmidt F, Weisblum Y et al (2021) mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants. Nature 592:616–622. https://doi.org/10.1038/s41586-021-03324-6
Planas D, Veyer D, Baidaliuk A et al (2021) Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature 596:276–280. https://doi.org/10.1038/s41586-021-03777-9
Karim SSA, Karim QA (2021) Omicron SARS-CoV-2 variant: a new chapter in the COVID-19 pandemic. Lancet 398:2126–2128. https://doi.org/10.1016/S0140-6736(21)02758-6
Schmidt F, Weisblum Y, Rutkowska M et al (2021) High genetic barrier to SARS-CoV-2 polyclonal neutralizing antibody escape. Nature 600:512–516. https://doi.org/10.1038/s41586-021-04005-0
Collie S, Champion J, Moultrie H et al (2021) Effectiveness of BNT162b2 vaccine against omicron variant in South Africa. N Engl J Med. https://doi.org/10.1056/NEJMc2119270
Nemet I, Kliker L, Lustig Y et al (2021) Third BNT162b2 vaccination neutralization of SARS-CoV-2 omicron infection. N Engl J Med. https://doi.org/10.1056/NEJMc2119358
Gruell H, Vanshylla K, Tober-Lau P et al (2021) mRNA booster immunization elicits potent neutralizing serum activity against the SARS-CoV-2 Omicron variant. Res Square. https://doi.org/10.21203/rs.3.rs-1168453/v1
Hoffmann M, Krüger N, Schulz S et al (2022) The omicron variant is highly resistant against antibodymediated neutralization—implications for control of the COVID-19 pandemic. Cell. https://doi.org/10.1016/j.cell.2021.12.032
Osterman A, Baldauf H-M, Eletreby M et al (2021) Evaluation of two rapid antigen tests to detect SARS-CoV-2 in a hospital setting. Med Microbiol Immunol 210:65–72. https://doi.org/10.1007/s00430-020-00698-8
Quick J (2020) nCoV-2019 sequencing protocol v3 (LoCost) V.3. In: protocols.io. https://protocols.io/view/ncov-2019-sequencing-protocol-v3-locost-bh42j8ye. Accessed 04 Jan 2022
Tyson JR, James P, Stoddart D et al (2020) Improvements to the ARTIC multiplex PCR method for SARS-CoV-2 genome sequencing using nanopore. BioRxiv. https://doi.org/10.1101/2020.09.04.283077
Quick J (2020) artic-ncov2019. https://github.com/joshquick/artic-ncov2019/blob/master/primer_schemes/nCoV-2019/V3/nCoV-2019.tsv. Accessed 4 Jan 2022
DNA Pipelines R&D, Farr B, Rajan D, et al (2020) COVID-19 ARTIC v3 Illumina library construction and sequencing protocol. In: protocols.io. https://www.protocols.io/view/covid-19-artic-v3-illumina-library-construction-an-bibtkann. Accessed 4 Jan 2022
Artic Network (2020) rampart. https://github.com/artic-network/rampart. Accessed 4 Jan 2022
Artic Network. https://artic.network/ncov-2019
O’Toole Á, Scher E, Underwood A et al (2021) Assignment of epidemiological lineages in an emerging pandemic using the pangolin tool. Virus Evol. https://doi.org/10.1093/ve/veab064
Classification of Omicron (B.1.1.529): SARS-CoV-2 Variant of Concern. https://www.who.int/news/item/26-11-2021-classification-of-omicron-(b.1.1.529)-sars-cov-2-variant-of-concern. Accessed 04 Jan 2022
European Centre for Disease Prevention and Contro (2021) Implications of the emergence and spread of the SARS-CoV-2 B.1.1. 529 variant of concern (Omicron) for the EU/EEA
Thermo Fisher Scientific Confirms Detection of SARS-CoV-2 in Samples Containing the Omicron Variant with its TaqPath COVID-19 Tests. In: MediaRoom. https://thermofisher.mediaroom.com/2021-11-29-Thermo-Fisher-Scientific-Confirms-Detection-of-SARS-CoV-2-in-Samples-Containing-the-Omicron-Variant-with-its-TaqPath-COVID-19-Tests. Accessed 4 Jan 2022
Muenchhoff M, Mairhofer H, Nitschko H, Grzimek-Koschewa N, Hoffmann D, Berger A, Rabenau H, Widera M, Ackermann N, Konrad R, Zange S, Graf A, Krebs S, Blum H, Sing A, Liebl B, Wolfel R, Ciesek S, Drosten C, Protzer U, Boehm S, Keppler OT (2020) Multicentre comparison of quantitative PCR-based assays to detect SARS-CoV-2, Germany, March 2020. Euro Surveill. https://doi.org/10.2807/1560-7917.ES.2020.25.24.2001057
Covid-19—TIB MOLBIOL. https://www.tib-molbiol.de/covid-19. Accessed 4 Jan 2022
Seegene Allplex SARS-CoV-2 Variants I Assay. https://www.seegene.com/assays/allplex_sars-cov-2_variants_i_assay. Accessed 4 Jan 2022
Seegene Allplex SARS-CoV-2 Variants II Assay. https://www.seegene.com/assays/allplex_sars-cov-2_variants_ii_assay. Accessed 4 Jan 2022
Cov-Lineages. https://cov-lineages.org/global_report_B.1.1.529.html. Accessed 4 Jan 2022
SARS-CoV-2 V4.1 update for Omicron variant. https://community.artic.network/t/sars-cov-2-v4-1-update-for-omicron-variant/342). Accessed 7 Jan 2022
Bull RA, Adikari TN, Ferguson JM et al (2020) Analytical validity of nanopore sequencing for rapid SARS-CoV-2 genome analysis. Nat Commun 11:6272. https://doi.org/10.1038/s41467-020-20075-6
Acknowledgements
We are grateful to members of the Diagnostic Laboratory of Virology at the Max von Pettenkofer Institute for support. We would like to thank the Bavarian SARS-CoV-Public Health Laboratory Epidemiology Team at the LGL for support.
Funding
Open Access funding enabled and organized by Projekt DEAL. This work was supported in part by the German BMBF initiative “NaFoUniMedCovid19” (01KX2021), subproject B-FAST (to O.T.K.), and by the Free State of Bavaria (research initiatives Bay-VOC (M.M., A.G., S.K., H.B., O.T.K.) and FOR-COVID (M.M., O.T.K.).
Author information
Authors and Affiliations
Contributions
OTK designed the study. CD, MM, HA, SB, HM, ST and SK performed experiments. CD, MM, AG, PRW, HM NG-K, HB and OTK analyzed data. CD and PRW assembled graphics. CD, MM and OTK wrote the manuscript. All authors discussed the results and commented on the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethics approval
The generation and publication of SARS-CoV-2 sequences and accompanying metadata were approved by the local ethics committee at the Medical Faculty of LMU München (No. 21-0740).
Additional information
Edited by Matthias J. Reddehase.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Dächert, C., Muenchhoff, M., Graf, A. et al. Rapid and sensitive identification of omicron by variant-specific PCR and nanopore sequencing: paradigm for diagnostics of emerging SARS-CoV-2 variants. Med Microbiol Immunol 211, 71–77 (2022). https://doi.org/10.1007/s00430-022-00728-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00430-022-00728-7