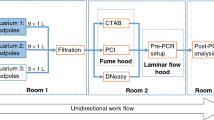
Abstract
Environmental DNA (eDNA) detection is a valuable conservation tool that can be used to identify and monitor imperiled or invasive species and wildlife pathogens. Batrachochytrium pathogens are of global conservation concern because they are a leading cause of amphibian decline. While eDNA techniques have been used to detect Batrachochytrium DNA in the environment, a systematic comparison of extraction methods across environmental samples is lacking. In this study, we first compared eDNA extraction methods and found that a soil extraction kit (Qiagen PowerSoil) was the most effective for detecting Batrachochytrium dendrobatidis in water samples. The PowerSoil extraction had a minimum detection level of 100 zoospores and had a two- to four-fold higher detection probability than other commonly used extraction methods (e.g., QIAamp extraction, DNeasy+Qiashredder extraction method, respectively). Next, we used this extraction method on field-collected water and sediment samples and were able to detect pathogen DNA in both. While field-collected water filters were equivalent to amphibian skin swab samples in detecting the presence of pathogen DNA, the seasonal patterns in pathogen quantity were different between skin swabs and water samples. Detection rate was lowest in sediment samples. We also found that detection probability increases with the volume of water filtered. Our results indicate that water filter eDNA samples can be accurate in detecting pathogen presence at the habitat scale but their utility for quantifying pathogen loads in the environment appears limited. We suggest that eDNA techniques be used for early warning detection to guide animal sampling efforts.




Similar content being viewed by others
Data availability
Data are available as 3 Online Resources files.
References
Audemard C, Reece KS, Eugene M, Burreson EM (2004) Real-time PCR for detection and quantification of the protistan parasite Perkinsus marinus in environmental waters. Appl Environ Microbiol 70:6611–6618. https://doi.org/10.1128/AEM.70.11.6611
Barnes MA, Brown AD, Daum MN et al (2020) Detection of the amphibian pathogens chytrid fungus (Batrachochytrium dendrobatidis) and ranavirus in West Texas, USA, using environmental DNA. J Wildl Dis 56:1–5. https://doi.org/10.7589/2019-08-212
Berger L, Speare R, Hyatt A (1999) Chytrid fungi and amphibian declines: Overview, implications and future directions. In: Campbell A (ed) Declines Disappearances Aust. Frogs, Environment Australia, Canberra, pp 23–33
Berger L, Speare R, Hines HB et al (2004) Effect of season and temperature on mortality in amphibians due to chytridiomycosis. Aust Vet J 82:434–439. https://doi.org/10.1111/j.1751-0813.2004.tb11137.x
Berger L, Roberts AA, Voyles J et al (2016) History and recent progress on chytridiomycosis in amphibians. Fungal Ecol 19:89–99. https://doi.org/10.1016/j.funeco.2015.09.007
Boyle DG, Boyle DB, Olsen V et al (2004) Rapid quantitative detection of chytridiomycosis (Batrachochytrium dendrobatidis) in amphibian samples using real-time Taqman PCR assay. Dis Aquat Organ 60:141–148. https://doi.org/10.3354/dao060141
Braid MD, Daniels LM, Kitts CL (2003) Removal of PCR inhibitors from soil DNA by chemical flocculation. J Microbiol Methods 52:389–393. https://doi.org/10.1016/S0167-7012(02)00210-5
Brannelly LA, Chatfield MWH, Richards-Zawacki CL (2012) Field and laboratory studies of the susceptibility of the green treefrog (Hyla cinerea) to Batrachochytrium dendrobatidis infection. PLoS ONE 7:e38473. https://doi.org/10.1371/journal.pone.0038473
Brannelly LA, Hunter DA, Lenger D et al (2015) Dynamics of chytridiomycosis during the breeding season in an Australian alpine amphibian. PLoS ONE 10:e0143629. https://doi.org/10.1371/journal.pone.0143629
Brannelly LA, Chatfield MWH, Sonn J et al (2018) Fungal infection has sublethal effects in a lowland subtropical amphibian population. BMC Ecol 18:34. https://doi.org/10.1186/s12898-018-0189-5
Brannelly L, Wetzel D, West M, Richards-Zawacki C (2020) Optimized swab sample extraction results in imperfect detection of Batrachochytrium dendrobatidis particularly when infection intensities are low. Dis Aquat Organ 139:233–243. https://doi.org/10.3354/dao03482
Chestnut T, Anderson C, Popa R et al (2014) Heterogeneous occupancy and density estimates of the pathogenic fungus Batrachochytrium dendrobatidis in waters of North America. PLoS ONE 9:e106790. https://doi.org/10.1371/journal.pone.0106790
Cossel JO Jr, Lindquist ED (2009) Batrachochytrium dendrobatidis in arboreal and lotic water sources in Panama. Herpetol Rev 40:45–47
Davies-Colley RJ, Hickey CW, Quinn JM, Ryan PA (1992) Effects of clay discharges on streams. Hydrobiologia 248:215–234
Dowd SE, Gerba CP, Pepper IL (1998) Confirmation of the human-pathogenic microsporidia Enterocytozoon bieneusi, Encephalitozoon intestinalis, and Vittaforma corneae in water. Appl Environ Microbiol 64:3332–3335
Goldberg CS, Turner CR, Deiner K et al (2016) Critical considerations for the application of environmental DNA methods to detect aquatic species. Methods Ecol Evol 7:1299–1307. https://doi.org/10.1111/2041-210X.12595
Hall EM, Crespi EJ, Goldberg CS (2015) Evaluating environmental DNA-based quantification of ranavirus infection in wood frog populations. Mol Ecol Resour 16:423–433. https://doi.org/10.1111/1755-0998.12461
Harper LR, Buxton AS, Rees HC et al (2019) Prospects and challenges of environmental DNA (eDNA) monitoring in freshwater ponds. Hydrobiologia 826:25–41. https://doi.org/10.1007/s10750-018-3750-5
Hot D, Legeay O, Jacques J et al (2003) Detection of somatic phages, infectious enteroviruses and enterovirus genomes as indicators of human enteric viral pollution in surface water. Water Res 37:4703–4710. https://doi.org/10.1016/S0043-1354(03)00439-1
Hyatt AD, Boyle DG, Olsen V et al (2007) Diagnostic assays and sampling protocols for the detection of Batrachochytrium dendrobatidis. Dis Aquat Organ 73:175–192. https://doi.org/10.3354/dao073175
Hyman OJ, Collins JP (2012) Evaluation of a filtration-based method for detecting Batrachochytrium dendrobatidis in natural bodies of water. Dis Aquat Organ 97:185–195. https://doi.org/10.3354/dao02423
Jerde CL, Mahon AR, Chadderton WL, Lodge DM (2011) “Sight-unseen” detection of rare aquatic species using environmental DNA. Conserv Lett 4:150–157. https://doi.org/10.1111/j.1755-263X.2010.00158.x
Kamoroff C, Goldberg C (2017) Using environmental DNA for early detection of amphibian chytrid fungus Batrachochytrium dendrobatidis prior to a ranid die-off. Dis Aquat Organ 127:75–79. https://doi.org/10.3354/dao03183
King HC, Murphy A, James P et al (2015) Performance of a noninvasive test for detecting Mycobacterium bovis shedding in European badger (Meles meles) populations. J Clin Microbiol 53:2316–2323. https://doi.org/10.1128/JCM.00762-15
Kirshtein JD, Anderson CW, Wood JS et al (2007) Quantitative PCR detection of Batrachochytrium dendrobatidis DNA from sediments and water. Dis Aquat Organ 77:11–15. https://doi.org/10.3354/dao01831
Kolby JE, Smith KM, Ramirez SD et al (2015) Rapid response to evaluate the presence of amphibian chytrid fungus (Batrachochytrium dendrobatidis) and ranavirus in wild amphibian populations in Madagascar. PLoS ONE 10:1–21. https://doi.org/10.1371/journal.pone.0125330
Kriger KM, Hero J-M, Ashton KJ (2006) Cost efficiency in the detection of chytridiomycosis using PCR assay. Dis Aquat Organ 71:149–154. https://doi.org/10.3354/dao071149
Lass A, Dumètre A, Szostakowska B, Myjak P (2009) Detection of Toxoplasma gondii oocysts in environmental soil samples using molecular methods. Eur J Clin Microbiol Infect Dis 28:599–605. https://doi.org/10.1007/s10096-008-0681-5
Lindner DL, Gargas A, Lorch JM et al (2011) DNA-based detection of the fungal pathogen Geomyces destructans in soils from bat hibernacula. Mycologia 103:241–246. https://doi.org/10.3852/10-262
Mo Bio Laboratories Inc (2014) PowerSoil® DNA isolation kit. Carlsbad, CA
Mosher BA, Huyvaert KP, Bailey LL (2018) Beyond the swab: ecosystem sampling to understand the persistence of an amphibian pathogen. Oecologia 188:319–330. https://doi.org/10.1007/s00442-018-4167-6
Nusser SM, Clark WR, Otis DL, Huang L (2008) Sampling considerations for disease surveillance in wildlife populations. J Wildl Manage 72:52–60. https://doi.org/10.2193/2007-317
Phillott AD, Grogan LF, Cashins SD et al (2013) Chytridiomycosis and seasonal mortality of tropical stream-associated frogs 15 years after introduction of Batrachochytrium dendrobatidis. Conserv Biol 27:1058–1068. https://doi.org/10.1111/cobi.12073
Pilliod DS, Goldberg CS, Arkle RS, Waits LP (2014) Factors influencing detection of eDNA from a stream-dwelling amphibian. Mol Ecol 14:109–116. https://doi.org/10.1111/1755-0998.12159
R Core Team (2017) R: a language and environmental for statistical computing. R Core Team, Vienna
Rees HC, Maddison BC, Middleditch DJ et al (2014) Review: The detection of aquatic animal species using environmental DNA—a review of eDNA as a survey tool in ecology. J Appl Ecol 51:1450–1459. https://doi.org/10.1111/1365-2664.12306
Richards-Zawacki CL (2010) Thermoregulatory behaviour affects prevalence of chytrid fungal infection in a wild population of Panamanian golden frogs. Proc R Soc Biol Sci 277:519–528. https://doi.org/10.1098/rspb.2009.1656
Sauer EL, Cohen JM, Lajeunesse MJ et al (2020) A meta-analysis reveals temperature, dose, life stage, and taxonomy influence host susceptibility to a fungal parasite. Ecology. https://doi.org/10.1002/ecy.2979
Scheele BC, Pasmans F, Skerratt LF et al (2019) Amphibian fungal panzootic causes catastrophic and ongoing loss of biodiversity. Science 363:1459–1463. https://doi.org/10.1126/science.aav0379
Schmidt BR, Kery M, Ursenbacher S et al (2013) Site occupancy models in the analysis of environmental DNA presence/absence surveys: a case study of an emerging amphibian pathogen. Methods Ecol Evol 4:646–653. https://doi.org/10.1111/2041-210X.12052
Shanan S, Abd H, Hedenström I et al (2011) Detection of Vibrio cholerae and Acanthamoeba species from same natural water samples collected from different cholera endemic areas in Sudan. BMC Res Notes 4:109. https://doi.org/10.1186/1756-0500-4-109
Smart AS, Tingley R, Weeks AR et al (2015) Environmental DNA sampling is more sensitive than a traditional survey technique for detecting an aquatic invader. Ecol Appl 25:1944–1952
Spitzen-van der Sluijs A, Stark T, DeJean T et al (2020) Using environmental DNA for detection of Batrachochytrium salamandrivorans in natural water. Environ DNA. https://doi.org/10.1002/edn3.86
Stegen G, Pasmans F, Schmidt BR et al (2017) Drivers of salamander extirpation mediated by Batrachochytrium salamandrivorans. Nature 544:353–358. https://doi.org/10.1038/nature22059
Thomsen PF, Willerslev E (2015) Environmental DNA—an emerging tool in conservation for monitoring past and present biodiversity. Biol Conserv 183:4–18. https://doi.org/10.1016/j.biocon.2014.11.019
Turner CR, Uy KL, Everhart RC (2015) Environmental DNA fish environmental DNA is more concentrated in aquatic sediments than surface water. Biol Conserv 183:93–102. https://doi.org/10.1016/j.biocon.2014.11.017
Walker SF, Salas MB, Jenkins D et al (2007) Environmental detection of Batrachochytrium dendrobatidis in a temperate climate. Dis Aquat Organ 77:105–112. https://doi.org/10.3354/dao01850
Acknowledgements
We thank our field assistants Phoebe Ruben, Trina Wantman, Jakub Zegar, Jeff Bednark, Chris Davis and Jessica Barabas. We thank Caren Goldberg, Emily Hall, Jon Kolby, and Sheri Sanders for advice processing the environmental samples.
Funding
This work was funded by the University of Pittsburgh’s Dietrich School of Arts and Sciences, the US National Science Foundation (IOS: Project No. 1649443) and the US Department of Defense (SERDP: Project No. RC-2638).
Author information
Authors and Affiliations
Contributions
LAB and CLRZ conceived and designed the experiment; LAB, MEBO, LZ, VS conducted the field component; LAB, LZ, DW conducted the molecular component; CLRZ contributed reagents and funded the experiment; LAB analyzed the data and wrote the manuscript; all authors contributed to editing the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Ethical approval
This research was conducted under the University of Pittsburgh Institutional Animal Care and Use Committee protocol 16027711. Animal surveys were conducted with permission from the Pennsylvania Fish and Boat Commission (permit 2017-01-0177), and from the Louisiana Department of Wildlife and Fisheries (permit LNHP-17-029).
Additional information
Communicated by Joel Trexler.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Brannelly, L.A., Wetzel, D.P., Ohmer, M.E.B. et al. Evaluating environmental DNA as a tool for detecting an amphibian pathogen using an optimized extraction method. Oecologia 194, 267–281 (2020). https://doi.org/10.1007/s00442-020-04743-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00442-020-04743-4