Abstract
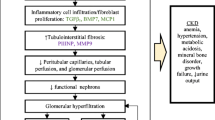
Chronic kidney disease (CKD) can progress to kidney failure and require dialysis or transplantation, while early diagnosis can alter the course of disease and lead to better outcomes in both pediatric and adult patients. Significant CKD comorbidities include the manifestation of cardiovascular disease, heart failure, coronary disease, and hypertension. The pathogenesis of chronic kidney diseases can present as subtle and especially difficult to distinguish between different glomerular pathologies. Early detection of adult and pediatric CKD and detailed mechanistic understanding of the kidney damage can be helpful in delaying or curtailing disease progression via precise intervention toward diagnosis and prognosis. Clinically, serum creatinine and albumin levels can be indicative of CKD, but often are a lagging indicator only significantly affected once kidney function has severely diminished. The evolution of proteomics and mass spectrometry technologies has begun to provide a powerful research tool in defining these mechanisms and identifying novel biomarkers of CKD. Many of the same challenges and advances in proteomics apply to adult and pediatric patient populations. Additionally, proteomic analysis of adult CKD patients can be transferred directly toward advancing our knowledge of pediatric CKD as well. In this review, we highlight applications of proteomics that have yielded such biomarkers as PLA2R, SEMA3B, and other markers of membranous nephropathy as well as KIM-1, MCP-1, and NGAL in lupus nephritis among other potential diagnostic and prognostic markers. The potential for improving the clinical toolkit toward better treatment of pediatric kidney diseases is significantly aided by current and future development of proteomic applications.

Similar content being viewed by others
References
United States Renal Data System (2019) 2019 USRDS annual data report: epidemiology of kidney disease in the United States. National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases, Bethesda, MD. https://www.usrds.org/annual-data-report/. Accessed 16 Aug 2021
Davis MT, Beierle J, Bures ET et al (2001) Automated LC-LC-MS-MS platform using binary ion-exchange and gradient reversed-phase chromatography for improved proteomic analyses. J Chromatogr B Biomed Sci App 752:281–291. https://doi.org/10.1016/s0378-4347(00)00547-8
Vissers JPC, Blackburn RK, Moseley MA (2002) A novel interface for variable flow nanoscale LC/MS/MS for improved proteome coverage. J Am Soc Mass Spectrom 13:760–771. https://doi.org/10.1016/S1044-0305(02)00418-X
Wolters DA, Washburn MP, Yates JR (2001) An automated multidimensional protein identification technology for shotgun proteomics. Anal Chem 73:5683–5690. https://doi.org/10.1021/ac010617e
McDonald WH, Yates JR (2002) Shotgun proteomics and biomarker discovery. Dis Markers 18:99–105. https://doi.org/10.1155/2002/505397
Dayon L, Hainard A, Licker V et al (2008) Relative quantification of proteins in human cerebrospinal fluids by MS/MS using 6-plex isobaric tags. Anal Chem 80:2921–2931. https://doi.org/10.1021/ac702422x
Wang Z, Yu K, Tan H et al (2020) 27-Plex tandem mass tag mass spectrometry for profiling brain proteome in Alzheimer’s disease. Anal Chem 92:7162–7170. https://doi.org/10.1021/acs.analchem.0c00655
Khurana M, Traum AZ, Aivado M et al (2006) Urine proteomic profiling of pediatric nephrotic syndrome. Pediatr Nephrol 21:1257–1265
Juraschek SP, Coresh J, Inker LA et al (2013) Comparison of serum concentrations of β-trace protein, β2-microglobulin, cystatin C, and creatinine in the US population. Clin J Am Soc Nephrol 8:584–592. https://doi.org/10.2215/CJN.08700812
Inker LA, Tighiouart H, Coresh J et al (2016) GFR Estimation Using β-Trace Protein and β2-Microglobulin in CKD. Am J Kidney Dis 67:40–48. https://doi.org/10.1053/j.ajkd.2015.07.025
Argyropoulos CP, Chen SS, Ng Y-H et al (2017) Rediscovering beta-2 microglobulin as a biomarker across the spectrum of kidney diseases. Front Med 4:73. https://doi.org/10.3389/fmed.2017.00073
Woroniecki RP, Shatat IF, Supe K et al (2008) Urinary cytokines and steroid responsiveness in idiopathic nephrotic syndrome of childhood. Am J Nephrol 28:83–90. https://doi.org/10.1159/000109396
Agrawal S, Merchant ML, Kino J et al (2020) Predicting and defining steroid resistance in pediatric nephrotic syndrome using plasma proteomics. Kidney Int Rep 5:66–80. https://doi.org/10.1016/j.ekir.2019.09.009
Al-Rabadi LF, Caza T, Trivin-Avillach C et al (2021) Serine protease HTRA1 as a novel target antigen in primary membranous nephropathy. J Am Soc Nephrol 32:1666–1681. https://doi.org/10.1681/ASN.2020101395
Tomas NM, Beck LH, Meyer-Schwesinger C et al (2014) Thrombospondin type-1 domain-containing 7A in idiopathic membranous nephropathy. N Engl J Med 371:2277–2287. https://doi.org/10.1056/NEJMoa1409354
Beck LH, Bonegio RGB, Lambeau G et al (2009) M-type phospholipase A2 receptor as target antigen in idiopathic membranous nephropathy. N Engl J Med 361:11–21. https://doi.org/10.1056/NEJMoa0810457
Sethi S, Debiec H, Madden B et al (2020) Semaphorin 3B-associated membranous nephropathy is a distinct type of disease predominantly present in pediatric patients. Kidney Int 98:1253–1264. https://doi.org/10.1016/j.kint.2020.05.030
Pérez V, López D, Boixadera E et al (2017) Comparative differential proteomic analysis of minimal change disease and focal segmental glomerulosclerosis. BMC Nephrol 18:49. https://doi.org/10.1186/s12882-017-0452-6
Choi YW, Kim YG, Song M-Y et al (2017) Potential urine proteomics biomarkers for primary nephrotic syndrome. Clin Proteomics 14:18. https://doi.org/10.1186/s12014-017-9153-1
Waikar SS, Sabbisetti VS, Bonventre JV (2010) Normalization of urinary biomarkers to creatinine during changes in glomerular filtration rate. Kidney Int 78:486–494. https://doi.org/10.1038/ki.2010.165
Suresh CP, Saha A, Kaur M et al (2016) Differentially expressed urinary biomarkers in children with idiopathic nephrotic syndrome. Clin Exp Nephrol 20:273–283. https://doi.org/10.1007/s10157-015-1162-7
Menon R, Otto EA, Hoover P et al (2020) Single cell transcriptomics identifies focal segmental glomerulosclerosis remission endothelial biomarker. JCI Insight 5:e133267. https://doi.org/10.1172/jci.insight.133267
Wenderfer SE, Gaut JP (2017) Glomerular diseases in children. Adv Chronic Kidney Dis 24:364–371. https://doi.org/10.1053/j.ackd.2017.09.005
Dotz V, Visconti A, Lomax-Browne H et al (2021) O- and N-glycosylation of serum immunoglobulin A is associated with IgA nephropathy and glomerular function. J Am Soc Nephrol 32:2455–2465. https://doi.org/10.1681/ASN.2020081208
Khositseth S, Kanitsap N, Warnnissorn N, Thongboonkerd V (2007) IgA nephropathy associated with Hodgkin’s disease in children: a case report, literature review and urinary proteome analysis. Pediatr Nephrol 22:541–546. https://doi.org/10.1007/s00467-006-0382-1
Fang X, Lu M, Xia Z et al (2021) Use of liquid chromatography-tandem mass spectrometry to perform urinary proteomic analysis of children with IgA nephropathy and Henoch-Schönlein purpura nephritis. J Proteomics 230:103979. https://doi.org/10.1016/j.jprot.2020.103979
Boneparth A, Wenderfer SE, Moorthy LN et al (2017) Clinical characteristics of children with membranous lupus nephritis: the Childhood Arthritis and Rheumatology Research Alliance Legacy Registry. Lupus 26:299–306. https://doi.org/10.1177/0961203316662720
Boneparth A, Ilowite NT, CARRA Registry Investigators (2014) Comparison of renal response parameters for juvenile membranous plus proliferative lupus nephritis versus isolated proliferative lupus nephritis: a cross-sectional analysis of the CARRA Registry. Lupus 23:898–904. https://doi.org/10.1177/0961203314531841
Pereira M, Muscal E, Eldin K et al (2017) Clinical presentation and outcomes of childhood-onset membranous lupus nephritis. Pediatr Nephrol 32:2283–2291. https://doi.org/10.1007/s00467-017-3743-z
Suzuki M, Wiers K, Brooks EB et al (2009) Initial validation of a novel protein biomarker panel for active pediatric lupus nephritis. Pediatr Res 65:530–536. https://doi.org/10.1203/PDR.0b013e31819e4305
Anania VG, Yu K, Pingitore F et al (2019) Discovery and qualification of candidate urinary biomarkers of disease activity in Lupus nephritis. J Proteome Res 18:1264–1277. https://doi.org/10.1021/acs.jproteome.8b00874
Traum AZ, Schachter AD (2007) Proteomic analysis in pediatric renal disease. Semin Nephrol 27:652–657. https://doi.org/10.1016/j.semnephrol.2007.09.009
Jo HA, Hyeon JS, Yang SH et al (2021) Fumarate modulates phospholipase A2 receptor autoimmunity-induced podocyte injury in membranous nephropathy. Kidney Int 99:443–455. https://doi.org/10.1016/j.kint.2020.06.031
Poyan Mehr A, Tran MT, Ralto KM et al (2018) De novo NAD+ biosynthetic impairment in acute kidney injury in humans. Nat Med 24:1351–1359. https://doi.org/10.1038/s41591-018-0138-z
Feng J, Zhang Q, Zhou Y et al (2018) Integration of proteomics and metabolomics revealed metabolite-protein networks in ACTH-secreting pituitary adenoma. Front Endocrinol 9:678. https://doi.org/10.3389/fendo.2018.00678
Bahado-Singh R, Poon LC, Yilmaz A et al (2017) Integrated proteomic and metabolomic prediction of term preeclampsia. Sci Rep 7:16189. https://doi.org/10.1038/s41598-017-15882-9
Costanzo M, Zacchia M, Bruno G et al (2017) Integration of proteomics and metabolomics in exploring genetic and rare metabolic diseases. Kidney Dis (Basel) 3:66–77. https://doi.org/10.1159/000477493
Cao H, Zhang A, Sun H et al (2015) Metabolomics-proteomics profiles delineate metabolic changes in kidney fibrosis disease. Proteomics 15:3699–3710. https://doi.org/10.1002/pmic.201500062
Sha Q, Lyu J, Zhao M et al (2020) Multi-omics analysis of diabetic nephropathy reveals potential new mechanisms and drug targets. Front Genet 11:616435. https://doi.org/10.3389/fgene.2020.616435
Pinu FR, Beale DJ, Paten AM et al (2019) Systems biology and multi-omics integration: viewpoints from the metabolomics research community. Metabolites 9:E76. https://doi.org/10.3390/metabo9040076
Chong J, Wishart DS, Xia J (2019) Using MetaboAnalyst 4.0 for comprehensive and integrative metabolomics data analysis. Curr Protoc Bioinforma 68:e86. https://doi.org/10.1002/cpbi.86
Pang Z, Chong J, Zhou G et al (2021) MetaboAnalyst 5.0: narrowing the gap between raw spectra and functional insights. Nucleic Acids Res 49:W388–W396. https://doi.org/10.1093/nar/gkab382
Wang B, Mezlini AM, Demir F et al (2014) Similarity network fusion for aggregating data types on a genomic scale. Nat Methods 11:333–337. https://doi.org/10.1038/nmeth.2810
Rohart F, Gautier B, Singh A, Lê Cao KA (2017) mixOmics: an R package for 'omics feature selection and multiple data integration. PLoS Comput Biol 13:e1005752
Argelaguet R, Velten B, Arnol D et al. (2018) Multi-omics factor analysis-a framework for unsupervised integration of multi-omics data sets. Mol Syst Biol 14:e8124. https://doi.org/10.15252/msb.20178124
Basu S, Duren W, Evans CR et al (2017) Sparse network modeling and metscape-based visualization methods for the analysis of large-scale metabolomics data. Bioinforma Oxf Engl 33:1545–1553. https://doi.org/10.1093/bioinformatics/btx012
Amezquita RA, Lun ATL, Becht E et al (2020) Orchestrating single-cell analysis with Bioconductor. Nat Methods 17:137–145. https://doi.org/10.1038/s41592-019-0654-x
Bioconductor. Open source software for bioinformatics. https://bioconductor.org/. Accessed 17 Aug 2021
Subramanian I, Verma S, Kumar S et al (2020) Multi-omics data integration, interpretation, and its application. Bioinforma Biol Insights 14:1177932219899051. https://doi.org/10.1177/1177932219899051
Reel PS, Reel S, Pearson E et al (2021) Using machine learning approaches for multi-omics data analysis: a review. Biotechnol Adv 49:107739. https://doi.org/10.1016/j.biotechadv.2021.107739
Francescatto M, Chierici M, Rezvan Dezfooli S et al (2018) Multi-omics integration for neuroblastoma clinical endpoint prediction. Biol Direct 13:5. https://doi.org/10.1186/s13062-018-0207-8
Sealfon RSG, Mariani LH, Kretzler M, Troyanskaya OG (2020) Machine learning, the kidney, and genotype-phenotype analysis. Kidney Int 97:1141–1149. https://doi.org/10.1016/j.kint.2020.02.028
Saez-Rodriguez J, Rinschen MM, Floege J, Kramann R (2019) Big science and big data in nephrology. Kidney Int 95:1326–1337. https://doi.org/10.1016/j.kint.2018.11.048
Bülow RD, Dimitrov D, Boor P, Saez-Rodriguez J (2021) How will artificial intelligence and bioinformatics change our understanding of IgA Nephropathy in the next decade? Semin Immunopathol 43:739–752. https://doi.org/10.1007/s00281-021-00847-y
Caza TN, Hassen SI, Kuperman M et al (2021) Neural cell adhesion molecule 1 is a novel autoantigen in membranous lupus nephritis. Kidney Int 100:171–181. https://doi.org/10.1016/j.kint.2020.09.016
Ayoub I, Shapiro JP, Song H et al (2021) Establishing a case for anti-complement therapy in membranous nephropathy. Kidney Int Rep 6:484–492. https://doi.org/10.1016/j.ekir.2020.11.032
Barwinska D Ferkowicz MJ Cheng YH Winfree S Dunn KW Kelly KJ Sutton TA Rovin BH Parikh SV Phillips CL Dagher PC El-Achkar TM Eadon MT; Kidney Precision Medicine Project (2020) application of laser microdissection to uncover regional transcriptomics in human kidney tissue. J Vis Exp 160:https://doi.org/10.3791/61371. https://doi.org/10.3791/61371
Hobeika L, Barati MT, Caster DJ et al (2017) Characterization of glomerular extracellular matrix by proteomic analysis of laser-captured microdissected glomeruli. Kidney Int 91:501–511. https://doi.org/10.1016/j.kint.2016.09.044
Lennon R, Byron A, Humphries JD et al (2014) Global analysis reveals the complexity of the human glomerular extracellular matrix. J Am Soc Nephrol 25:939–951. https://doi.org/10.1681/ASN.2013030233
Merchant ML, Barati MT, Caster DJ et al (2020) Proteomic analysis identifies distinct glomerular extracellular matrix in collapsing focal segmental glomerulosclerosis. J Am Soc Nephrol 31:1883–1904. https://doi.org/10.1681/ASN.2019070696
Clotet-Freixas S, McEvoy CM, Batruch I et al (2020) Extracellular matrix injury of kidney allografts in antibody-mediated rejection: a proteomics study. J Am Soc Nephrol 31:2705–2724. https://doi.org/10.1681/ASN.2020030286
Paunas FTI, Finne K, Leh S et al (2019) Characterization of glomerular extracellular matrix in IgA nephropathy by proteomic analysis of laser-captured microdissected glomeruli. BMC Nephrol 20:410. https://doi.org/10.1186/s12882-019-1598-1
Solier C, Langen H (2014) Antibody-based proteomics and biomarker research - current status and limitations. Proteomics 14:774–783. https://doi.org/10.1002/pmic.201300334
Cody EM, Bennett MR, Gulati G et al (2021) Successful urine multiplex bead assay to measure lupus nephritis activity. Kidney Int Rep 6:1949–1960. https://doi.org/10.1016/j.ekir.2021.04.016
Niewczas MA, Pavkov ME, Skupien J et al (2019) A signature of circulating inflammatory proteins and development of end-stage renal disease in diabetes. Nat Med 25:805–813. https://doi.org/10.1038/s41591-019-0415-5
Potter SS, Brunskill EW (2014) Building an atlas of gene expression driving kidney development: pushing the limits of resolution. Pediatr Nephrol 29:581–588. https://doi.org/10.1007/s00467-013-2602-9
Funding
This work was supported by NIDDK-NIH (DK110077 to JBK, WES), and clinical research funding from the University of Louisville Kidney Disease Program.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
William E. Smoyer and Jon B. Klein are co-senior authors
Rights and permissions
About this article
Cite this article
Cummins, T.D., Korte, E.A., Bhayana, S. et al. Advances in proteomic profiling of pediatric kidney diseases. Pediatr Nephrol 37, 2255–2265 (2022). https://doi.org/10.1007/s00467-022-05497-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00467-022-05497-2