Abstract
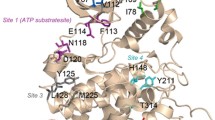
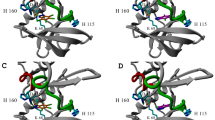
Protein kinase CK2 (casein kinase 2) is a multifunctional serine/threonine kinase that is involved in a broad range of physiological events. The decreased affinity of Emodin binding to human CK2α resulting from single-point mutation of Val66 to Ala (V66A) has been demonstrated by experimental mutagenesis. Molecular dynamics (MD) simulations and energy analysis were performed on wild type (WT) and V66A mutant CK2α-Emodin complexes to investigate the subtle influences of amino acid replacement on the structure of the complex. The structure of CK2α and the orientation of Emodin undergo changes to different degrees in V66A mutant. The affected positions in CK2α are mainly distributed over the glycine-rich loop (G-loop), the α-helix and the loop located at the portion between G-loop and α-helix (C-loop). Based on the coupling among these segments, an allosteric mechanism among the C-loop, the G-loop and the deviated Emodin is proposed. Additionally, an estimated energy calculation and residue-based energy decomposition also indicate the lower instability of V66A mutant in contrast to WT, as well as the unfavorable energetic influences on critical residue contributions.








Similar content being viewed by others
References
Litchfield DW (2003) Protein kinase CK2: structure, regulation and role in ellular decisions of life and death. Biochem J 369:1–15
Duncan JS, Litchfield DW (2008) Too much of a good thing: The role of protein kinase CK2 in tumorigenesis and prospects for the therapeutic inhibition of CK2. Biochim Biophys Acta 1784:33–47
Meggio F, Pinna LA (2003) One-thousand-and one substrates of protein kinase CK2? FASEB J 17:349–368
Ahmad KA, Wang G, Slaton J, Unger G, Ahmed K (2005) Targeting CK2 for cancer therapy. Anticancer Drugs 16:1037–1043
Unger GM, Davis AT, Slaton JW, Ahmed K (2004) Protein kinase CK2 as regulator of cell survival: implications for cancer therapy. Curr Cancer Drug Targets 4:77–84
Bibby AC, Litchfield DW (2005) The multiple personalities of the regulatory subunit of protein kinase CK2: CK2 dependent and CK2 independent roles reveal a secret identity for CK2 beta. Int J Biol Sci 1:67–79
Pinna LA (2002) Protein kinase CK2: a challenge to canons. J Cell Sci 115:3873–3878
Mazzorana M, Pinna LA, Battistutta R (2008) A structural insight into CK2 inhibiton. Mol Cell Biochem 316:57–62
Prudent R, Cochet C (2009) New protein kinase CK2 inhibitors: Jumping out of the catalytic box. Chem Biol 16:112–120
Niefind K, Putter M, Guerra B, Issinger OG, Schomburg D (1999) GTP plus water mimic ATP in the active site of protein kinase CK2. Nat Struct Bio 6:1100–1103
Battistutta R, Sarno S, De Moliner E, Papinutto E, Zanotti G, Pinna LA (2000) The replacement of ATP by the competitive inhibitor Emodin induces conformational modifications in the catalytic site of Protein kinase CK2. J Bio Chem 275:29618–29622
Sarno S, Sali M, Battistutta R, Zanotti G, Pinna LA (2005) Features and potentials of ATP-site directed CK2 inhibitors. Biochim Biophys Acta 1754:263–270
Battistutta R, De Moliner E, Sarno S, Zanotti G, Pinna LA (2001) Structural features underlying selective inhibition of protein kinase CK2 by ATP site-directed tetrabromo-2-benzotriazole. Protein Sci 10:2200–2206
De Moliner E, Moro S, Sarno S, Zagotto G, Zanotti G, Pinna LA, Battistutta R (2003) Inhibition of protein kianse CK2 by Anthraquinone-related compounds. J Biol Chem 278:1831–1836
Battistutta R, Mazzorana M, Cendron L, Bortolato A, Sarno S, Kazimierczuk Z, Zanotti G, Moro S, Pinna LA (2007) The ATP-binding site of protein kinase CK2 holds a positive electrostatic area and conserved water molecules. Chembiochem 8:1804–1809
Battistutta R, Mazzorana M, Sarno S, Kazimierczuk Z, Zanotti G, Pinna LA (2005) Inspecting the structure-activity relationship of protein kinase CK2 inhibitors derived from tetrabromo-benzimidazole. Chem Biol 12:1211–1219
Chilin A, Battistutta R, Bortolato A, Cozza G, Zanatta S, Poletto G, Mazzorana M, Zagotto G, Uriarte E, Guiotto A, Pinna LA, Meggio F, Moro S (2008) Coumarin as attractive casein kinase 2 (CK2)inhibitor scaffold: an integrate approach to elucidate the putativebinding motif and explain structure-activity relationships. J Med Chem 51:752–759
Sarno S, Moro S, Meggio F, Zagotto G, Dal Ben D, Ghiselline P, Battistutta R, Zanotti G, Pinna LA (2002) Toward the rational design of protein kinase casein kinase-2 inhibitors. Pharmacol Ther 93:159–168
Pagano MA, Bain J, Kazimierczuk Z, Sarno Stefania S, Ruzzene M, Di Maira G, Elliott M, Orzeszko A, Cozza G, Meggio F, Pinna LA (2008) The selectivity of inhibitors of protein kinase CK2: an update. Biochem J 415:353–365
Sarno S, Vaglio P, Meggio F, Ruzzene M, Davies SP, Donella Deana A, Shugar D, Pinna LA (2001) Selectivity of 4, 5, 6, 7-terabromobenzotriazole, an ATP-site-directed inhibitor of protein kinase CK2 (‘casein kinase-2’). FEBS Lett 496:44–48
Zhang N, Jiang YJ, Zou JW, Zhuang SL, Jin HX, Yu QS (2007) Insights into unbinding mechanisms upon two mutations investigated by molecular dynamics study of GSK3β-Axin complex: Role of packing hydrophobic residues. Proteins 67:941–949
Zhang N, Jiang YJ, Zou JW, Zhao WN, Yu QS (2009) Structural basis for the complete loss of GSK3β catalytic activity due to R96 mutation investigated by molecular dynamics study. Proteins 75:671–681
Gohlke H, Case DA (2004) Converging free energy estimates: MM-PB(GB)SA studies on the protein-protein complex Ras-Raf. J Comput Chem 25:238–250
LepšíK M, Kříž Z, Havlas Z (2004) Efficiency of a second generation HIV-1 protease inhibitor studied by molecular dynamics and absolute binding free energy calculations. Proteins 57:279–293
Raaf J, Klopffleisch K, Issinger O-G, Niefind K (2008) The catalytic subunit of Human protein kinase CK2 structurally deviates from Its maize homologue in complex with the nucleotide competitive inhibitor Emodin. J Mol Biol 377:1–8
Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Zakrzewski VG, Montgomery JA Jr, Stratmann RE, Burant JC, Dapprich S, Millam JM, Daniels AD, Kudin KN, Strain MC, Farkas O, Tomasi J, Barone V, Cossi M, Cammi R, Mennucci B, Pomelli C, Adamo C, Clifford S, Ochterski J, Petersson GA, Ayala PY, Cui Q, Morokuma K, Malick DK, Rabuck AD, Raghavachari K, Foresman JB, Cioslowski J, Ortiz JV, Stefanov BB, Liu G, Liashenko A, Piskorz P, Komaromi I, Gomperts R, Martin RL, Fox DJ, Keith T, Al-Laham MA, Peng CY, Nanayakkara A, Gonzalez C, Challacombe M, Gill PMW, Johnson BG, Chen W, Wong MW, Andres JL, Head-Gordon M, Replogle ES, Pople JA (2003) Gaussian 03, Revision C.02. Gaussian Inc, Wallingford, CT
Besler BH, Merz KM, Kollman PA (1990) Atomic charges derived from semiempirical methods. J Comput Chem 11:431–439
Fox T, Kollman PA (1998) Application of the RESP methodology in the parametrization of organic solvents. J Phys Chem B 102:8070–8079
Case DA, Darden T, Cheathem TE III, Simmerling C, Wang JM, Duke RE, Luo R, Merz KM, Wang B, Pearlman DA, Croley M, Brozell S, Tsui V, Gohleke H, Mongan J, Hornak V, Cui GL, Beroza P, Schafmeister C, Caldwell JW, Ross WS, Kollman PA (2008) AMBER 10. University of California, San Francisco
Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML (1983) Comparison of simple potential functions for simulating liquid water. J Chem Phys 79:926–935
Duan Y, Wu C, Chowdhury S, Lee MC, Xiong G, Zhang W, Yang R, Cieplak P, Luo R, Lee T (2003) A point-charge force field for molecular mechanics simulations of proteins. J Comput Chem 24:1999–2012
Berendsen HJC, Postma JPM, van Gunsteren WF, Di Nola A, Haak JR (1984) Molecular dynamics with coupling to an external bath. J Chem Phys 81:3684–3690
Ryckaert JP, Ciccotti G, Berendsen HJC (1977) Numerical integration of the cartesian equations of motion of a system with constraints:molecular dynamics of n-alkanes. J Comput Phys 23:327–341
Darden T, York D, Pedersen L (1993) Particle mesh Ewald: an N log (N) method for Ewald sums in large systems. J Chem Phys 98:10089–10094
Massova I, Kollman PA (2000) Combined molecular mechanical and continuum solvent approach (MM-PBSA/GBSA) to predict ligand binding. Persp Drug Discov Des 18:113–135
Kollman PA, Massova I, Reyes C, Kuhn B, Shuanghong H, Chong L, Lee M, Lee T, Duan Y, Wang W, Donini O, Cieplak P, Srinivasan J, Case DA, Cheatham TE III (2000) Calculating structures and free energies of complex molecules: Combining molecular mechanics and continuum models. Acc Chem Res 33:889–897
Sitkoff D, Sharp KA, Honig B (1994) Accurate calculation of hydration free energies using macroscopic solvent models. J Phys Chem 98:1978–1988
Wang W, Kollman PA (2001) Computational study of protein specificity: The molecular basis of HIV-1 protease drug resistance. Proc Natl Acad Sci USA 98:14937–14942
Wang J, Morin P, Wang W, Kollman PA (2001) Use of MM-PBSA in reproducing the binding free energies to HIV-1 RT of TIBO derivatives and predicting the binding mode to HIV-1 RT of efavirenz by docking and MM-PBSA. J Am Chem Soc 123:5221–5230
Jayaram B, Sprous D, Beveridge DL (1998) Solvation free energy of biomacromolecules: parameters for a modified generalized born model consistent with the AMBER force field. J Phys Chem B 102:9571–9576
Gohlke H, Kiel C, Case DA (2003) Insights into protein-protein binding by binding free energy calculation and free energy decomposition for the Ras-Raf and Ras-RalGDS complexes. J Mol Biol 330:891–913
Welburn JP, Tucker JA, Johnson T, Lindert L, Morgan M, Willis A, Nobe MEM, Endicott JA (2007) How Tyrosine 15 phosphorylation inhibits the activity of Cyclin-dependent kinase 2-cyclin A. J Biol Chem 282:3173–3181
Aimes RT, Hemmer W, Taylor SS (2000) Serine-53 at the tip of the glycine-rich loop of cAMP-dependent protein kinase: role in catalysis, P-site specificity, and interaction with inhibitors. Biochem 39:8325–8332
Grant B, Hemmer W, Tsigelny I, Adams JA, Taylor SS (1998) Kinetic analyses of mutations in the glycine-rich loop of cAMP-dependent protein kinase. Biochemistry 37:7708–7715
Sun H, Jiang YJ, Yu QS, Luo CC, Zou JW (2008) Effect of mutation K85R on GSK-3β: Molecular dynamics simulation. Biochem Biophys Res Commun 377:962–965
Jin HX, Wu TX, Jiang YJ, Zou JW, Zhuang SL, Mao X, Yu QS (2007) Role of phosphorylated Thr-197 in the catalytic subunit of cAMP-dependent protein kinase. J Mol Struct 805:9–15
Schindler T, Bornmann W, Pellicena P, Miller WT, Clarkson B, Kuriyan J (2000) Structural mechanism for STI-571 inhibition of abelson tyrosine kinase. Science 289:1938–1942
Mohammadi M, McMahon G, Sun L, Tang C, Hirth P, Yeh BK, Hubbar SR, Schlessinger J (1997) Structures of the tyrosine kinase domain of fibroblastgrowth factor receptor in complex with inhibitors. Science 276:955–960
Niefind K, Yde CW, Ermakova I, Issinger OG (2007) Evolved to be active: Sulfate ions define substrate recognition sites of CK2α and emphasise its exceptional role within the CMGC family of eukaryotic protein kinases. J Mol Biol 370:427–438
Yde CW, Ermakova I, Niefind K (2005) Incling the purine base binding plane in protein kinase CK2 by exchanging the flanking side-chains generates a preference for ATP as a cosubstrate. J Mol Biol. 347:399–414
Raaf J, Issinger OG, Niefind K (2009) First inactive conformation of CK2α, the catalytic subunit of protein kinase CK2. J Mol Biol 386:1212–1221
De Moliner E, Brown NR, Johnson LN (2003) Alternative binding modes of an inhibitor to two different kinases. Eur J Biochem 270:3174–3181
Acknowledgments
This work was funded by China Postdoctoral Science Foundation funded project (No. 20090450271) and Beijing Natural Science Foundation (8072006). We acknowledge Professor David Case for the kind gift of AMBER 10 software.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Zhang, N., Zhong, R. Structural basis for decreased affinity of Emodin binding to Val66-mutated human CK2α as determined by molecular dynamics. J Mol Model 16, 771–780 (2010). https://doi.org/10.1007/s00894-009-0582-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00894-009-0582-2