Abstract
Charge sensitivity analysis (CSA) in force-field atoms resolution was applied to describe the mutual polarization of reactants as well as charge-transfer (CT) effects. An inclusion complex of β-cyclodextrin with salicylic acid was used as a model system. Three CSA models were taken into account and verified on a Born–Oppenheimer molecular dynamics (BOMD) trajectory. The models differed in terms of the equilibrium conditions imposed on the system. It was demonstrated that mutual polarization is an important source of stabilization, in contrast to the results obtained from static charge calculations. The energy lowering induced by CT was small and comparable to the CT stabilization that occurs in hydrogen-bonded systems. All models correctly described the main topological features of the BOMD energy surface. CSA in force-field atoms resolution qualitatively reproduced the charge reorganization accompanying hydrogen-bond formation. It was shown that CSA parameters are very sensitive to the bond formation process, which suggests that they could be applied in reactive force fields as detectors of newly formed chemical bonds.

Fukui function detector of bond formation during molecular dynamics simulations of inclusion complex of b-cyclodextrin with salicylic acid.
Similar content being viewed by others
Introduction
Classical molecular mechanics (MM) and molecular dynamics (MD) simulations have become powerful tools for studying large molecular systems. They can deal with systems consisting of hundreds of thousands of atoms, and can simulate their dynamic behavior over a time scale of nanoseconds. They are especially important when studying biomolecules and condensed phases, as the results of such simulations complement experimental results and provide insight into microscopic properties that cannot be probed experimentally [1, 2]. They are capable of effectively treating these large-scale systems through the utilization of simplified functions that express the system’s potential energy U(R) in terms of the coordinates of the atoms, \( {\bf{R}} = \left( {{{{\bf{R}}}_{1}},\:{{{\bf{R}}}_{2}}, \ldots ,\:{{{\bf{R}}}_{N}}} \right) \), where N is the total number of atoms. In the most popular additive force fields [3–9], electrostatic contributions to U(R) are described using the Coulomb potential between static atomic partial charges:
where q i and q j are the charges on atoms i and j, respectively, ε is the dielectric constant, and \( {{{\bf{R}}}_{{ij}}} = \left| {{{{\bf{R}}}_{i}} - {{{\bf{R}}}_{j}}} \right| \) is the distance between atoms i and j.
In additive force fields, many-body effects—particularly polarization—are neglected (i.e., electronic degrees of freedom are ignored, and the charge distribution is assumed to remain constant during the simulation). To implicitly account for the polarization that occurs in condensed phases, partial atomic charges determined for the gas phase are systematically overestimated by 10–20 % [10, 11]. This approach takes into account the potential energy contributions neglected in U(R) using an average. There are, however, a variety of systems for which such an approximation is insufficient. In general, these systems contain inhomogeneities where polarizable molecules experience changes in the dielectric environment, for example in interfacial regions: liquid–vapor, liquid–liquid, or protein–solvent surfaces, or solutions of ions. Polarization also plays an important role when various thermodynamic processes have to be considered.
Currently models that explicitly include polarization in MD simulations include polarizable dipoles [12–14], the classical Drude oscillator [15, 16], and the fluctuating charge (FQ) model [17–23]. The first two models allow for the occurrence of induced dipole moments at various sites in the system (atoms, bonds, lone pairs). In the FQ model, the values of atomic partial charges are allowed to vary. This is based on the electronegativity equalization (EE) principle [24–26]. The Drude and FQ models have the advantage of preserving the Coulombic potential for electrostatic interactions without introducing an additional dipole–dipole term. In MD simulations, all three models can be solved iteratively or with extended Lagrangian mechanics [27]. The FQ model can also be solved using matrix inversion.
Charge sensitivity analysis (CSA) was first formulated in the 1990s in the framework of density functional theory (DFT) [28, 29]. Although it was originally designed as a tool to complement ab initio calculations, it can—after suitable parameterization—be used as a semi-empirical method. It provides information about the charge distribution in a molecular system and about the system’s reactivity (Fukui function and softness/hardness data). It is based on the second-order Taylor expansion of the system’s energy with respect to population variables and on the EE principle. From this point of view, the FQ model is similar to CSA. However, this is only one aspect of CSA. CSA undoubtedly provides a wider perspective. The reactivity information built into the CSA formalism can be used in qualitative structure–activity relationship (QSAR) and qualitative structure–property relationship (QSPR) models. The huge potential of conceptual DFT in such models has been demonstrated in the literature [30–36]. In our previous paper [37], we parameterized CSA for the AMBER ff99 [38, 39] force field on the basis of static calculations performed in the global equilibrium state. In this paper, we present the utilization of CSA for other types of equilibria. In MD simulations, it is typical to employ a reactant perspective—each reactant preserves its own identity, manifested as a unique set of bonding parameters. In this paper, in addition to global equilibrium, we will use the approach of imposing a partial equilibrium on the system (i.e., the reactants are mutually polarized but there is no charge transfer between them). Moreover, we will explore the dynamic behavior of partial charges and other CSA parameters along a simulated trajectory of a molecular system.
The paper is organized as follows. We first present a short survey of CSA, before introducing a model system and providing computational details. We then describe the obtained results, and finally present our conclusions and future prospects in this field.
Charge sensitivity analysis
The basis of CSA is the Taylor expansion of the energy of a molecular system M with respect to atomic charges, truncated at the quadratic term:
Here, q is the vector grouping atomic partial charges, \( {{\mathbf{q}}^{\dag}}=\left( {{q_1},\;{q_2},\ldots,\;{q_N}} \right) \), \( \boldsymbol{\upchi}_0^{\dag}=\left( {\chi_1^0,\;\chi_2^0,\ldots,\;\chi_N^0} \right)=\left\{ {\chi_i^0={{{\partial {E_{\text{M}}}}} \left/ {{\partial {q_i}}} \right.}} \right\} \) is the vector of atomic electronegativities, \( {{\boldsymbol{\upeta}}_0}=\left\{ {\eta_{ij}^0={{{{\partial^2}{E_{\text{M}}}}} \left/ {{\partial {q_i}\partial {q_j}}} \right.}} \right\} \) is the hardness matrix, and N is the total number of atoms in the system. An index of “0” indicates that derivatives were evaluated for q = 0. Electronegativity represents the ability of an atom to attract electrons and, in the framework of DFT, is identified as the negative chemical potential [40]. It fulfills Sanderson’s electronegativity equalization principle, which states that for the equilibrium charge distribution, electronegativity is equalized throughout space:
In other words, electrons flow between atoms until the atomic electronegativities are equalized. Note that the electronegativity in an open system (global equilibrium) is equal to \( {\chi_i}=\chi_i^0+\sum\nolimits_j {\eta_{ij}^0{q_j}} \). In MD simulations, one often deals with molecular systems constructed from smaller subsystems (e.g., individual molecules). It is convenient to impose a charge conservation constraint on each of these subsystems. Electronegativity is then equalized in each of the fragments but not in the system as a whole.
By taking the derivative of Eq. 2 with respect to the i-th atomic charge and applying the EE principle (Eq. 3), one arrives at a set of linear equations for the equilibrium charge distribution in M. If M is constructed from two subsystems A and B, M = (A|B) (the solid line indicates no charge transfer between A and B), the CSA equations can be summarized as the following single matrix equation:
where 1 A (1 B) and 0 A (0 B) are unity and zero vectors of dimension N A (N B), and \( \chi_{\text{A}}^{\text{eq} } \) and \( \chi_{\text{B}}^{\text{eq} } \) are the fragments’ equalized electronegativities. The hardness matrix η 0 and the vector that groups the atomic electronegativities χ 0 are divided into sub-blocks. The first two rows in Eq. 4 represent the charge conservation constraints for A and B on q A and q B, respectively. The remaining equations describe electronegativity equalization.
The equilibrium charge distribution in M is obtained by inverting the matrix in Eq. 4:
The elements of the inverse matrix are: diagonal Fukui function (FF) indices, \( {{\mathbf{f}}_{{\text{AA}} }}=\left\{ {{{{\partial {q_{{i\in {\text{A}}}}}}} \left/ {{\partial {q_{\text{A}}}}} \right.}} \right\} \) and \( {{\mathbf{f}}_{{\text{BB}} }}=\left\{ {{{{\partial {q_{{i\in {\text{B}}}}}}} \left/ {{\partial {q_{\text{B}}}}} \right.}} \right\} \); off-diagonal Fukui function indices, \( {{\mathbf{f}}_{{\text{AB}} }}=\left\{ {{{{\partial {q_{{i\in {\text{A}}}}}}} \left/ {{\partial {q_{\text{B}}}}} \right.}} \right\} \) and \( {{\mathbf{f}}_{{\text{BA}} }}=\left\{ {{{{\partial {q_{{i\in {\text{B}}}}}}} \left/ {{\partial {q_{\text{A}}}}} \right.}} \right\} \); the condensed hardness matrix \( {{\boldsymbol{\upeta}}_{{\text{frg}} }}=\left\{ {{{{{\eta_{XY }}=\partial {\chi_X}}} \left/ {{\partial {q_Y},}} \right.}\;X,\,\,Y=\left( {{\text{A}},\;{\text{B}}} \right)} \right\} \), and the polarization matrix \( {{\boldsymbol{\upbeta}}_{XY }}=\left\{ {{{{\partial {q_{{i\in X}}}}} \left/ {{\partial {v_{{j\in Y}}}}} \right.}} \right\}, \) \( X,\,Y=\left. {\left( {{\text{A}},\;{\text{B}}} \right)} \right\} \). The Fukui function describes the response of a given atom to a perturbation in the total number of electrons in its fragment (diagonal FF) or another fragment (off-diagonal FF). Diagonal FF vectors are normalized to unity, f AA 1 A † = f BB 1 B † = 1, while off-diagonal FF vectors are normalized to zero, f AB 1 B † = f BA 1 A † = 0. The block (β XY ) of the polarization matrix represents the charge reorganization within X induced by perturbation in the external potential on the j-th atom of Y (v j∈Y ). Both the diagonal and off-diagonal blocks of the polarization matrix are normalized to zero:
Atomic (diagonal) hardnesses describe an atom’s strength to block electron flow. They can be related to the hard and soft acids and bases (HSAB) principle [41]. In the frozen core approximation, the off-diagonal elements of the hardness matrix can be related to the Coulomb exchange integrals between valence shell s electrons:
They can be further approximated by neglecting the exchange integrals, K ij , and reducing η ij to the Coulomb integrals J ij . Therefore, one can determine the crudest approximation of \( \eta_{{ij}}^0 \) as 1/R ij . However, for short separations, the utilization of 1/R ij results in incorrect behavior, and semi-empirical combination formulae should be used instead. In accordance with our previous studies [42], we adopted Ohno’s formula [43]:
where \( {a_{ij }}={2 \left/ {{\left( {\eta_{ii}^0+\eta_{jj}^0} \right)}} \right.}\equiv {2 \left/ {{\left( {\eta_i^0+\eta_j^0} \right)}} \right.} \). Therefore, only diagonal hardnesses are needed to construct the hardness matrix.
The empirical parameters of CSA are the effective electronegativities and diagonal hardnesses of atoms in molecules. These depend on the atomic number, hybridization, and chemical environment of a given atom, and are determined by reproducing the gas-phase charge distribution calculated with an ab initio method for a set of training molecules. In our previous work [37, 42], we determined CSA parameters based on Mulliken population analysis (MPA) [44] and a hybridization/chemical environment classification inherited from the AMBER ff99 force field [38].
Model systems and computational details
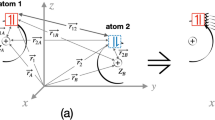
An inclusion complex of β-cyclodextrin (BCD) with salicylic acid (SAL) was used to illustrate the performance of CSA. The BCD molecule contains seven glucopyranose residues which form a shallow truncated cone with primary and secondary hydroxyl groups exposed to the solvent. The primary hydroxyl groups form the smaller rim of the cone. The secondary hydroxyl groups form the bigger rim. The molecule contains a hydrophobic cavity and is able to incorporate many organic molecules. Inclusion complexes of BCD with different “guest” molecules are stabilized by weak interactions (van der Waals and hydrogen bonds). At the classical MD level of theory, each reactant preserves its identity, so complexes of BCD are good candidates to use when describing mutual polarization. In addition, the guest molecule breaks the high symmetry of the host (BCD) molecule. This leads to huge charge distribution inhomogeneities in glucopyranose residues and allows us to check the parameterization of CSA in reactant resolution.
BCD has several conformers; more information about their structures can be found in [45–49]. The conformer with a counterclockwise orientation of hydrogen bonds in both rims was chosen. The guest molecule was assumed to be in a planar conformation. The most stable conformer of SAL contains an intramolecular hydrogen bond between the phenyl hydroxyl hydrogen atom and the carbonyl oxygen atom from the carboxylic group. A less stable conformer was chosen for the MD calculation since, at first glance, it appears better suited to inter-reactant hydrogen-bond formation. The structures of the host and guest molecules are shown in Scheme 1.
The initial structures of both molecules were obtained at the B3LYP/6-31G(d) level of theory using the Gaussian 09 [50] suite of programs. The starting geometry of the complex was obtained by placing the SAL molecule on the C7 symmetry axis of BCD, with the line joining the C1 and C4 aromatic carbons coinciding with the symmetry axis. This initial structure was the starting point in Born–Oppenheimer MD (BOMD) calculations. The TeraChem [51] general purpose quantum chemistry software package was used. A 10 ps MD run with 1 fs time steps at 300 K was performed at the B3LYP/6-31G level of theory. An empirical dispersion correction was utilized. The trajectory obtained was recalculated at the molecular mechanics (MM) level of theory. The CHARMM force field for hydrocarbons was employed. Data concerning pyranose monosaccharides and glycoside linkages were taken from [52] and [53]. In our previous work [37], we parameterized CSA using a diverse training set consisting of small organic molecules and AMBER ff99 all-atom force-field resolution. In the CHARMM force field, however, the sp 3 carbon atoms are additionally differentiated into secondary, primary, tertiary, and quaternary, as well as according to their chemical environment. None of these features were observed when force-field resolution was achieved with AMBER ff99. The classification of aliphatic hydrogen atoms also differed between the two force fields. In order to preserve the consistency of the CHARMM carbohydrate force field, we corrected the CSA parameters for BCD using the same protocol as before and a training set containing mono- and polyhydroxylated alcohols, simple linear and cyclic ethers, carbohydrates (aldoses, ketoses, furanoses, pyranoses, and disaccharides), and other model compounds. For the SAL molecule, defined using the CHARMM general force field for small molecules, the atom types were consistent with the previous CSA resolution, so the original parameter set was retained. All parameters are collected in Table 1. Three types of MM runs were performed using the NAMD package [54]. In the first MM run, the charge distribution was frozen. The Mulliken charges obtained at the B3LYP/6-31G(d) level were used. In the second and third MM runs, charges were derived using Eq. 5 for each point on the trajectory. In the second MM run, the charges in each subsystem were constrained (see Eq. 4). In the third run, CT between subsystems was allowed, and only the global charge of the BCD/SAL complex was constrained. In other words, two first rows/columns of Eq. 4 were replaced with one row/column: (0, 1 N ). These three types of calculations are termed the electrostatic (ES), polarization (P), and charge-transfer (CT) runs throughout the paper; these names reflect the physical effects that are additionally included in the MM calculations. The ES, P, and CT runs are characterized by the following equilibria: \( {{\text{M}}^{{\text{ES}} }}=\left( {\,{1_{\text{A}}}\left| {\,{2_{\text{A}}}} \right.\left| {\,\ldots } \right.\left| {\,{N_{\text{A}}}} \right.\left| {\,{1_{\text{B}}}\left| {\,{2_{\text{B}}}} \right.\left| {\,\ldots } \right.\left| {\,{N_{\text{B}}}} \right.} \right.} \right) \), \( {{\text{M}}^{\text{P}}}=\left( {\,{1_{\text{A}}}\vdots\,{2_{\text{A}}}\vdots\,\ldots \vdots\,{N_{\text{A}}}\left| {\,{1_{\text{B}}}\vdots\,{2_{\text{B}}}\vdots\,\ldots \vdots\,{N_{\text{B}}}} \right.} \right) \), and \( {{\text{M}}^{{\text{CT}} }}=\left( {{1_{\text{A}}}\vdots\,{2_{\text{A}}}\vdots\,\ldots \vdots\,{N_{\text{A}}}\vdots {1_{\text{B}}}\vdots\,{2_{\text{B}}}\vdots\,\ldots \vdots\,{N_{\text{B}}}} \right) \). Solid/dotted lines indicate that charge flow between atoms is forbidden/allowed.
Results and discussion
A detailed analysis of the trajectory obtained allowed us to distinguish the following steps in the inclusion process: (i) the O7′ and H7′ atoms of SAL form a hydrogen bond with the O6 atom of BCD (inter-reactant hydrogen bond); (ii) the O2 and H2 atoms of SAL form a hydrogen bond with O2 of BCD (inter-reactant hydrogen bond); (iii) the O2 and H2 atoms of SAL form a hydrogen bond with O3 of BCD (inter-reactant hydrogen bond); (iv) O2, H2, and O7 of SAL form a hydrogen bond (intra-reactant hydrogen bond). Steps (ii) and (iii) have intermediate character. These inter-reactant hydrogen bonds appear to consecutively disappear after a conformational change in the SAL molecule and the formation of the intra-reactant hydrogen bond (step (iv)). The hydrogen bonds formed in steps (i) and (iv) are preserved for the rest of the simulation. The first is formed for t ∈ (700,900), while the second is formed for t ∈ (6400, 6600). Therefore, two main domains can be distinguished: one for t ∈ (900, 6400) and the other for t ∈ (6600, 10000).
Energy and charge profiles
Figure 1 shows the time evolution of the system’s energy. Note that the B3LYP/6-31G total energy (red curve) was shifted by a constant value in order to allow all of the curves to be plotted together. In general, the remaining curves should be shifted, since none of the force-field terms resulting from Taylor expansions include the zero-order contribution (e.g., bond stretching term). Two main domains can be distinguished along the trajectory. The average energy in the first domain (after step (i)) is higher than that in the second domain (after step (iv)). The relative positions of the curves on the energy scale agree with expectation: mutual polarization introduces additional stabilization into the system. It can be seen that the P (purple) and CT (blue, dotted line) curves are shifted down by about 100 kcal/mol with respect to the ES (green) curve. By removing the barrier to charge flow between BCD and the guest molecule, further stabilization is introduced. The CT stabilization doesn’t exceed 0.5 kcal/mol, so the P and CT curves are practically indistinguishable. Such energy lowering is typical in the hydrogen-bonded systems [55–57].
The fluctuations in BOMD energy are smaller than those seen in the MM runs (ES, P, and CT). The MM curves differ only in the electrostatic contribution. The geometries at a given time step are exactly the same for all runs. Therefore, the bonding and van der Waals energies are also the same. This indicates that the bigger amplitudes seen for the P, CT, and ES runs are due to the bonding and van der Waals terms, which can be corrected for by adjusting some of the force-field parameters.
The shapes of ES, P, and CT curves are almost identical, and reproduce the main topological features of the BOMD curve. Figures S1 and S2 of the “Electronic supplementary material” (ESM) clearly demonstrate this, as they present the first and second derivatives of the system’s energy with respect to time for a narrow region of the trajectory, 6400 ≤ t ≤ 6600. This part of the trajectory was chosen since it corresponds to the hydrogen-bond formation that restores the most stable conformer of the SAL molecule (step (iv)). It is evident that the BOMD energy profile is very well reproduced. The positions of the extrema and the curvatures of all of the curves are almost identical.
A diagram showing the correlation between the B3LYP/6-31 and CSA charges is shown in Fig. 2. This figure includes all atoms for t ∈ (6400, 6600). The correlation coefficient is higher than 0.99. Our charges are slightly underestimated with respect to the B3LYP charges. Some of these discrepancies are due to basis set inconsistency: namely, the CSA model was parameterized for 6-31G(d) charges while, due to a lack of d-functions in the TeraChem package, they were compared with the 6-31G charges. The correlation diagram shows the average characteristic. It is important to determine whether the trends in charge reorganization accompanying hydrogen-bond formation are the same in both sets. Figures S3a and S3b of the ESM relate to the MPA and CSA charges, respectively. Charges from the CT run and the P run are indistinguishable, so only one of these sets (that from the CT run) was plotted. The main features of MPA charge evolution are preserved by CSA. Namely, O7 and H2 (the hydroxyl hydrogen atom) exhibit “step-like” population changes.
CSA parameter profiles
Inclusion in the BCD cavity can modify the physicochemical properties of the guest molecules (solubility, diffusion, sublimation, volatility, chromatographic mobility, hydrophobicity, reactivity, etc.) [58]. The influence of BCD on the physicochemical properties of the guest molecule can be seen in Figs. 3 and 4, which show electronegativity and hardness data. Figure 3 presents the reactant electronegativities χ SAL (green curve) and χ BCD (red curve), together with the global electronegativity χ (blue curve). The most important observation that can be made from this figure is the fact that the electronegativity of the whole inclusion complex is almost the same as the electronegativity of the BCD molecule. A similar observation follows from Fig. 4. The hardness of the inclusion complex η (global hardness) is the same as the diagonal hardness η BCD,BCD. Such behavior demonstrates that the electronic properties of the inclusion complex are closer to those of the host BCD molecule. In other words, the presence of the guest molecule is masked and the SAL/BCD complex resembles the host molecule. Taking into account the bioadaptability of BCD [59], it is not surprising that BCD has found widespread application as an encapsulating material for medicaments and food ingredients, since the weak interactions stabilizing the complex allows the guest molecule to be released at the target location.
The history of the inclusion process is reflected in the CSA data. The parameters most sensitive to bond formation are the polarization matrix elements and the diagonal FF indices. The diagonal FF indices and polarization matrix elements correspond to the P run [intra-reactant equilibrium: \( {{{\rm{M}}}^{{\rm{P}}}} = ({{1}_{{\rm{A}}}} \vdots \,{{2}_{{\rm{A}}}} \vdots \, \ldots \vdots \,{{N}_{{\rm{A}}}}\left| {{{1}_{{\rm{B}}}} \vdots \,{{2}_{{\rm{B}}}} \vdots \, \ldots \vdots \,{{N}_{{\rm{B}}}}} \right.) \)]. One can expect that the atoms involved in hydrogen-bond formation are the most sensitive to a perturbation in the external potential or in the number of electrons.
In Fig. 5, the population responses of the hydrogen-accepting oxygen to an external potential perturbation on the hydrogen atom (black curve) and the hydrogen-donating oxygen atom (red curve) are plotted. The polarization matrix is symmetric, so these elements also describe the population responses of the hydrogen and hydrogen-donating oxygen atoms to an external potential perturbation on the hydrogen-accepting oxygen atom (Maxwell relations). Parts A and B correspond to inter-reactant (step (i)) and intra-reactant (step (iv)) hydrogen bonds, respectively. One can observe step-like behavior by \( \beta_{\mathrm{H}7\text{',}\mathrm{O}6}^{\mathrm{SAL}\text{,}\mathrm{BCD}} \), \( \beta_{\mathrm{O}7\text{',}\mathrm{O}6}^{\mathrm{SAL}\text{,}\mathrm{BCD}} \) (part A) and \( \beta_{\mathrm{H}\text{2,}\mathrm{O}7}^{\mathrm{SAL}\text{,}\mathrm{SAL}} \), \( \beta_{\mathrm{O}\text{2,}\mathrm{O}7}^{\mathrm{SAL}\text{,}\mathrm{SAL}} \) (part B). The jump in the average value of \( \beta_{\mathrm{H}7\text{',}\mathrm{O}6}^{\mathrm{SAL}\text{,}\mathrm{BCD}} \) or \( \beta_{\mathrm{H}\text{2,}\mathrm{O}7}^{\mathrm{SAL}\text{,}\mathrm{SAL}} \) is greater than that of \( \beta_{\mathrm{O}7\text{',}\mathrm{O}6}^{\mathrm{SAL}\text{,}\mathrm{BCD}} \) or \( \beta_{\mathrm{O}\text{2,}\mathrm{O}7}^{\mathrm{SAL}\text{,}\mathrm{SAL}} \).
Time evolutions of selected elements of the polarization matrix. Part A presents \( \beta_{\mathrm{H}7\text{',}\mathrm{O}6}^{\mathrm{SAL}\text{,}\mathrm{BCD}} \) and \( \beta_{\mathrm{O}7\text{',}\mathrm{O}6}^{\mathrm{SAL}\text{,}\mathrm{BCD}}, \) while part B presents \( \beta_{\mathrm{H}\text{2,}\mathrm{O}7}^{\mathrm{SAL}\text{,}\mathrm{SAL}} \) and \( \beta_{\mathrm{O}\text{2,}\mathrm{O}7}^{\mathrm{SAL}\text{,}\mathrm{SAL}} \) (see Scheme 1 for atom labels)
Figure 6 presents the evolutions of diagonal FF indices over time. Atoms involved in the formation of the intra-fragment hydrogen bond (namely O2, H2, and O7 of SAL) were taken into account. The diagonal FF index f i AA= ∂q i A/∂q A, measures the response of the i-th atom population of A (SAL) to the oxidation/reduction of A as a whole. One can observe step-like behavior in all of the curves (in an averaged sense). This behavior is especially apparent for H2. After hydrogen-bond formation, the atom becomes harder and the initial average FF value of 0.10 is halved to 0.05 (FF can be considered as the normalized softness and hardness is the inverse of the softness). For the remaining atoms, the jump in FF index is less pronounced. The hydrogen-donating atom (O2) becomes harder after bond formation, while the hydrogen-accepting oxygen (O7) becomes softer. The variations in the off-diagonal FF indices are shown in Fig. 7. The same atoms involved in the intra-fragment hydrogen bond of the SAL molecule are considered. Off-diagonal effects are rather subtle in comparison with those for the diagonal FF indices. This is not surprising, since they measure the response of the i-th atom of SAL to the oxidation/reduction of BCD. None of the curves show step-like behavior. The values of \( f_{\mathrm{O}2}^{\mathrm{SAL}\text{,}\mathrm{BCD}} \) and \( f_{\mathrm{O}7}^{\mathrm{SAL}\text{,}\mathrm{BCD}} \) gradually increase from one domain (its average value) to the other domain. \( f_{\mathrm{H}2}^{\mathrm{SAL}\text{,}\mathrm{BCD}} \) exhibits huge fluctuations that are damped after intra-fragment hydrogen-bond formation. These fluctuations were connected with inter-reactant hydrogen-bond formation (steps (ii) and (iii)). The off-diagonal FF indices carry the “chemical” information (hybridization, rehybridization, polarization, etc.), especially in local or molecular orbital resolution [60, 61].
Time evolutions of the diagonal FF indices of the O2 (cyan), H2 (black), and O7 (green) atoms for  partitioning of the system, where A = SAL and B = BCD (see Scheme 1 for atom labels)
partitioning of the system, where A = SAL and B = BCD (see Scheme 1 for atom labels)
Time evolutions of the off-diagonal FF indices of the O2 (cyan), H2 (black), and O7 (green) atoms for  partitioning of the system, where A = SAL and B = BCD (see Scheme 1 for atom labels)
partitioning of the system, where A = SAL and B = BCD (see Scheme 1 for atom labels)
The clear and sharp character of the changes in FF indices and polarization matrix elements indicates that they should be a very good descriptors for detecting bond formation during MD simulations. In particular, they are better indicators than atomic charges, which have a smoother transition between the two domains. Of course, atomic charges cannot be used as indicators in static force fields.
Conclusions and future prospects
In this paper, polarization and charge-transfer contributions were incorporated into MM calculations using a CSA approach. CSA is based on electronegativity equalization equations supplemented by charge conservation equations. The results obtained for the BCD/SAL inclusion complex indicate that our model properly describes the energetics and charge reorganization that accompany the insertion of a guest molecule into the BCD cavity. In addition, the reactivity descriptors built into the CSA formalism can be used as indicators of specific chemical processes that occur in the system. This method can be easily extended to systems comprising a huge number of molecules. Work aiming at incorporating CSA in MD packages is currently being carried out. It appears that CSA parameters can be applied to developed reactive force fields which allow to investigate chemical reactions.
References
Mackerell AD Jr (2004) Empirical force fields for biological macromolecules: overview and issues. J Comput Chem 25:1584–1604
Schettino V, Chelli R, Marsili S, Barducci A, Faralli C, Pagliai M, Procacci P, Cardini G (2007) Problems in molecular dynamics of condensed phases. Theor Chem Accounts 117:1105–1120
Duan Y, Wu C, Chowdhury S, Lee MC, Xiong G, Zhang W, Yang R, Cieplak P, Luo R, Lee T, Caldwell JW, Wang J, Kollman PA (2003) A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase mechanical calculations. J Comput Chem 24:1999–2012
MacKerell AD Jr, Karplus M, Bashford D, Bellott M, Dunbrack RL Jr, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, Joseph-McCarthy D, Kuchnir L, Kuczera K, Lau FTK, Mattos C, Michnick S, Ngo T, Nguyen DT, Prodhom B, Reicher WE III, Roux B, Schlenkrich M, Smith JC, Stote R, Straub J, Watanabe M, Wiórkiewicz-Kuczera J, Yin D (1998) All-atom empirical potential for molecular modeling and dynamics studies of proteins. J Phys Chem B 102:3586–3616
van Gunsteren WF, Scott WRP, Hunenberger PH, Tironi IG, Mark AE, Billeter SR, Fennen J, Torda AE, Huber T, Kruger P (1999) The GROMOS biomolecular simulation program package. J Phys Chem A 103:3596–3607
Jorgensen WL, Tirado-Rives J (1988) The OPLS potential functions for proteins. Energy minimizations for crystals of cyclic peptides and crambin. J Am Chem Soc 110:1658–1666
Ewig CS, Berry R, Dinur U, Hill JR, Hwang MJ, Li H, Liang C, Maple JON, Peng Z, Stockfisch TP, Thatcher TS, Yan L, Ni X, Hagler AT (2001) Derivation of class II force fields. VIII. Derivation of a general quantum mechanical force field for organic compounds. J Comput Chem 22:1782–1800
Goddard WA III, Mayo SL, Olafson BD (1990) DREIDING: a generic force field for molecular simulations. J Phys Chem 94:8897–8909
Rappe AK, Casewit CJ, Colwell KS, Goddard WA III, Skiff WM (1992) UFF, a full periodic table force field for molecular mechanics and molecular dynamics simulations. J Am Chem Soc 114:10024–10035
Ponder JW, Case DA (2003) Force fields for protein simulations. Adv Protein Chem 66:27–85
Becker OM, Mackerell AD Jr, Roux B, Watanabe M (2001) Computational biochemistry and biophysics. Marcel Dekker Inc., New York
Sprik M, Klein ML (1988) A polarizable model for water using distributed charge sites. J Chem Phys 89:7556–7560
Liu YP, Kim K, Berne BJ, Friesner RA, Rick SW (1998) Constructing ab initio force fields for molecular dynamics simulations. J Chem Phys 108:4739–4755
Kaminski GA, Stern HA, Berne BJ, Friesner RA, Cao YX, Murphy RB, Zhou R, Halgren TA (2002) Development of a polarizable force field for proteins via ab initio quantum chemistry: first generation model and gas phase tests. J Comput Chem 23:1515–1531
Roux B, Lamoureux G (2003) Modeling induced polarization with classical drude oscillators: theory and molecular dynamics simulation algorithm. J Chem Phys 119:3025–3039
Mackerell AD, Lopes PEM, Lamoureux G (2008) Polarizable empirical force field for nitrogen-containing heteroaromatic compounds based on the classical drude oscillator. J Comput Chem 30:1821–1838
Mortier WJ, Ghosh SK, Shankar S (1986) Electronegativity equalization method for the calculation of atomic charges in molecules. J Am Chem Soc 108:4315–4320
Goddard WA III, Rappe AK (1991) Charge equlibration for molecular dynamics simulations. J Phys Chem 95:3358–3363
Rick SW, Stuart SJ, Bader JS, Berne BJ (1995) Fluctuating charge force fields for aqueous solutions. J Mol Liq 5:31–40
Rick SW, Berne BJ (1996) Dynamical fluctuating charge force fields: the aqueous solvation of amides. J Am Chem Soc 118:672–679
Rick SW (1997) Free energy of the hydrophobic interaction from molecular dynamics simulations: the effects of solute and solvent polarizability. J Phys Chem B 101:10488–10493
Ribeiro MCC, Almeida LCJ (1999) Fluctuating charge model for polyatomic ionic systems: a test case with diatomic anions. J Chem Phys 110:11445–11448
Llanta E, Ando K, Rey R (2001) Fluctuating charge study of polarization effects in chlorinated organic liquids. J Phys Chem B 105:7783–7791
Sanderson RT (1976) Chemical bond and bond energy. Academic Press, New York
Mortier WJ (1987) Electronegativity equalization and its applications. Struct Bond 66:125–143
Koca J, Jirouskova Z, Svobodova Varekova R, Vanek J (2009) Electronegativity equalization method: parameterization and validation for organic molecules using the Merz–Kollman–Singh charge distribution scheme. J Comput Chem 30:1174–1178
van Belle D, Lippens G, Froeyen M, Wodak SJ (1992) Molecular dynamics simulation of polarizable water by an extended Lagrangian method. Mol Phys 77:239–255
Nalewajski RF (2000) Charge sensitivities of the externally interacting open reactants. Int J Quantum Chem 78:168–178
Nalewajski RF, Korchowiec J (1998) Charge sensitivity approach to electronic structure and chemical reactivity. World Scientific Press, Singapore
Geerlings P, De Proft F, Langenaeker W (2003) Conceptual density functional theory. Chem Rev 103:1793–1873
Geerlings P, Ayers PW, Toro-Labbé A, Chattaraj PK, De Proft F (2012) The Woodward–Hoffmann rules reinterpreted by conceptual density functional theory. Acc Chem Res 45:683–695
Torrent-Sucarrat M, De Proft F, Ayers PW, Geerlings P (2010) On the applicability of local softness and hardness. Phys Chem Chem Phys 12:1072–1080
Sablon N, De Proft F, Solà M, Geerlings P (2012) The linear response kernel of conceptual DFT as a measure of aromaticity. Phys Chem Chem Phys 14:3960–3967
Ayers PW, Parr RG (2000) A theoretical perspective on the bond length rule of Grochala, Albrecht and Hoffmann. J Phys Chem A 104:2211–2220
Parr RG, Chattaraj PK, Cedillo A (1995) Variational method for determining the Fukui function and chemical hardness of an electronic system. J Chem Phys 103:7645–7646
Gupta K, Giri S, Chattaraj PK (2013) Charge-based DFT descriptors for Diels–Alder reactions. J Phys Org Chem. doi:10.1002/poc.2987
Stachowicz A, Korchowiec J (2012) Generalized charge sensitivity analysis. J Struct Chem 23:1449–1458
Kollman PA, Cornell WD, Cieplak P, Bayly CI, Gould IR, Merz KM Jr, Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW (1995) A second generation force field for the simulation of proteins, nucleic acids, and organic molecules. J Am Chem Soc 117:5179–5197
Cieplak P, Wang J, Kollman PA (2000) How well does a restrained electrostatic potential (RESP) model perform in calculating conformational energies of organic and biological molecules? J Comput Chem 21:1049–1074
Parr RG, Yang W (1989) Density functional theory of atoms and molecules. Oxford University Press, Oxford
Yang W, Parr RG (1985) Hardness, softness, and the Fukui function in the electronic theory of metals and catalysis. Proc Natl Acad Sci USA 82:6723–6726
Stachowicz A, Styrcz A, Korchowiec J (2011) Charge sensitivity analysis in force-field-atom resolution. J Mol Model 17:2217–2226
Ohno K (1964) Some remarks on the Pariser–Parr–Pople method. Theor Chem Accounts 2:219–227
Mulliken RS (1955) Electronic population analysis on LCAO-MO molecular wave functions. I. J Chem Phys 23:1833–1840
Britto MAFO, Nascimento CS, Hélio F (2004) Structural analysis of cyclodextrins: a comperative study of classical and quantum mechanical methods. Quím Nova 27:882–888
Heine T, dos Santos HF, Patchkovskii S, Duarte HA (2007) Structure and dynamics of beta-cyclodextrin in aqueous solution at the density-functional tight binding level. J Phys Chem A 111:5648–5654
Pinjari RV, Joshi KA, Gejji SP (2007) Theoretical studies on hydrogen bonding, NMR chemical shifts and electron density topography in alpha, beta and gamma-cyclodextrin conformers. J Phys Chem A 111:13583–13589
Snor W, Liedl E, Weiss-Greiler P, Karpfen A, Viernstein H, Wolschann P (2007) On the structure of anhydrous β-cyclodextrin. Chem Phys Lett 441:159–162
Korchowiec J, Stachowicz A, Styrcz A, Modaressi A, Rogalski M (2011) DFT studies of cation binding by β-cyclodextrin. Theor Chem Accounts 130:939–953
Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA et al (2009) Gaussian 09, Revision A.02. Gaussian Inc., Wallingford
Ufimtsev IS, Martinez TJ (2009) Quantum chemistry on graphical processing units. 3. Analytical energy gradients, geometry optimization, and first principles molecular dynamics. J Chem Theory Comput 5:2619–2628
Guvench O, Greene SN, Kamath G, Brady JW, Venable RM, Pastor RW, Mackerell AD Jr (2008) Additive empirical force field for hexopyranose monosaccharides. J Comput Chem 29:2543–2564
Guvench O, Hatcher ER, Venable RM, Pastor RW, Mackerell AD Jr (2009) CHARMM additive all-atom force field for glycosidic linkages between hexopyranoses. J Chem Theory Comput 5:2353–2370
Schulten K, Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kalé L (2005) Scalable molecular dynamics with NAMD. J Comput Chem 26:1781–1802
Korchowiec J, Uchimaru T (2000) New energy partitioning scheme based on the self-consistent charge and configuration method for subsystems: application to water dimer system. J Chem Phys 112:1623–1633
Uchimaru T, Korchowiec J, Tsuzuki S, Matsumura K, Kawahara S (2000) Importance of secondary electrostatic interactions in hydrogen-bonding complexes: an investigation using the self-consistent charge and configuration method for subsystems. Chem Phys Lett 318:203–209
de Silva P, Korchowiec J (2010) Energy partitioning scheme based on self-consistent method for subsystems: populational space approach. J Comput Chem 32:1054–1064
Szejtli J (1998) Introduction and general overview of cyclodextrin chemistry. Chem Rev 98:1743–1754
Loftsson T, Duchêne D (2007) Cyclodextrins and their pharmaceutical applications. Int J Pharm 329:1–11
Korchowiec J, Uchimaru T (1998) The charge transfer Fukui function: extension of the finite-difference approach to reactive systems. J Phys Chem A 102:10167–10172
Korchowiec J, Chandra AK, Uchimaru T, Kawahara S, Matsumura K, Tsuzuki S, Mikami M (1999) The mutual polarization of reactants: Fukui function description of the charge reorganization. Chem Phys Lett 308:229–234
Acknowledgments
CSA and MM calculations were performed on a computer cluster purchased with financial support from the European Regional Development Fund in the framework of the Polish Innovation Economy Operational Program (contract no. POIG.02.01.00-12-023/08). TeraChem calculations were performed at Akademickie Centrum Komputerowe (ACK) CYFRONET. The authors acknowledge financial support from the National Science Center (project no. UMO-2011/01/B/ST4/00636).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Fig. S1
Time evolution of the first derivative of the system’s energy with respect to time for a selected fragment of the simulation trajectory. Red curve indicates the BOMD run, green indicates the ES run, purple indicates the P run, and blue indicates the CT run (JPEG 271 kb)
Fig. S2
Time evolution of the second derivative of the system’s energy with respect to time for a selected fragment of the simulation trajectory. Red curve indicates the BOMD run, green indicates the ES run, purple indicates the P run, and blue indicates the CT run (JPEG 267 kb)
Fig. S3
Variation in charge versus time for selected SAL atoms, as calculated with MPA (part A) and CSA (part B). The atoms belong to the carboxyl group (red, green, blue and purple curves) and the hydroxyl group (black and cyan curves) (JPEG 469 kb)
(JPEG 465 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
About this article
Cite this article
Stachowicz, A., Rogalski, M. & Korchowiec, J. Charge sensitivity approach to mutual polarization of reactants: molecular mechanics perspective. J Mol Model 19, 4163–4172 (2013). https://doi.org/10.1007/s00894-013-1757-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00894-013-1757-4