Abstract
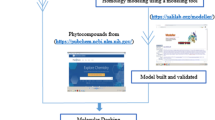
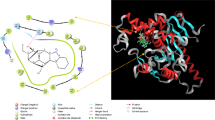
The cell wall of Mycobacterium tuberculosis interacts with the host counterpart during the pathogenesis of tuberculosis. L-rhamnosyl (L-Rha) residue, a linker connects the arabinogalactan and peptidoglycan moieties in the bacterial cell wall. The biosynthesis of L-rhamnose utilizes four successive enzymes RmlA, RmlB, RmlC and RmlD. Neither rhamnose nor the genes responsible for its synthesis are observed in humans. Thus, drugs inhibiting enzymes of this pathway are unlikely to interfere with metabolic pathways in humans. The adverse drug effects of first and second line drugs along with the development of multi-drug resistance tuberculosis have stimulated the research in search of new therapeutic drugs. Thus, it is attractive to hypothesize that inhibition of the biosynthesis of L-Rha would be lethal to the mycobacteria. Nature provides innumerable secondary metabolites with novel structural architectures with reported activity against M. tuberculosis. Combination of structure based virtual screening with physicochemical and pharmacokinetic studies against rhamnose pathway enzymes identified potential leads. The crucial screening studies recognized four phytocompounds butein, diospyrin, indicanine, and rumexneposide A with good binding affinity towards the rhamnose pathway proteins. Furthermore, the high throughput screening methods recognized butein, a secondary metabolite from Butea monosperma with strong anti-tubercular bioactive spectrum. Butein displayed promising anti-mycobacterial activity which is validated by Microplate alamar blue assay (MABA). The focus on novel agents like these phytocompounds which exhibit preference toward the successive enzymes of a single pathway can prevent the development of bacterial resistance.









Similar content being viewed by others
References
WHO global Tuberculosis report 2013
Louw GE, Warren RM, Geyvan NC, McEvoy CR, Van PD, Victor TC (2009) A balancing act: efflux/influx in mycobacterial drug resistance. Antimicrob Agents Chemother 53:3181–3189
Barun M, Natalia EK, Pablo JB, Barry NK (2006) Molecular epidemiology of tuberculosis: current insights. Clin Microbiol Rev 196:58–685
Stephen HG (2002) Evolution of drug resistance in mycobacterium tuberculosis: clinical and molecular perspective. Antimicrob Agents Chemother 46:267–274
Shanmugam A, Jeyakumar N (2012) Multi-targeted therapy for leprosy: insilico strategy to overcome multi drug resistance and to improve therapeutic efficiency. Infect Genet Evol 12:1899–1910
Ma Y, Stern RJ, Scherman MS, Vissa VD, Yan W, Jones VC et al. (2001) Drug targeting mycobacterium tuberculosis cell wall synthesis: genetics of dTDP-rhamnose synthetic enzymes and development of a microtiter plate-based screen for inhibitors of conversion of dTDP-glucose to dTDP-rhamnose. Antimicrob Agents Chemother 45:1407–1416
Yufang M, Fei P, Michael M (2002) Formation of dTDP-rhamnose is essential for growth of mycobacteria. J Bacteriol 184:3392–3395
Qu H, Xin Y, Dong X, Ma Y (2007) An rmlA gene encodingD-glucose-1-phosphate thymidylyltransferase is essential for mycobacterial growth. FEMS Microbiol Lett 275:237–243
Wulf B, Miryam A, Joseph SL, James HN (2000) The structural basis of the catalytic mechanism and regulation of glucose-1-phosphate thymidylyltransferase (RmlA). EMBO J 19:6652–6663
Allard ST, Giraud MF, Whitfield C, Graninger M, Messner P, Naismith JH (2001) The crystal structure of dTDP-D-glucose 4, 6-dehydratase (RmlB) from Salmonella enterica Serovar Typhimurium, the second enzyme in the dTDP-L-rhamnose pathway. J Mol Biol 307:283–295
Kantardjieff KA, Kim CY, Naranjo C, Waldo GS, Lekin T, Segelke BW, Zemla A, Park MS, Terwilliger TC, Rupp B (2004) Mycobacterium tuberculosis RmlC epimerase (Rv3465): a promising drug-target structure in the rhamnose pathway. Acta Crystallogr D Biol Crystallogr D60:895–902
Blankenfeldt W, Kerr ID, Giraud MF, McMiken HJ, Leonard G, Whitfield C, Messner P, Graninger M, Naismith JH (2002) Variation on a theme of SDR: dTDP-6-deoxy-L-lyxo-4-hexulose reductase (RmlD) shows a new Mg2+−dependent dimerization mode. Structure 10:773–786
Salomon CE, Schmidt LE (2012) Natural products as leads for tuberculosis drug development. Curr Top Med Chem 12:735–765
The UniProt Consortium (2013) Update on activities at the Universal Protein Resource (UniProt) in 2013. Nucleic Acids Res 41:D43–D47
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H et al. (2007) ClustalW and ClustalW version 2. Bioinformatics 23:2947–2948
Eswar N, Webb B, Marti Renom MA, Madhusudhan MS, Eramian D, Shen MY et al. (2007) Comparative protein structure modeling with MODELLER. Curr Protoc Protein Sci Chapter 2: Unit 2.9
Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H et al. (2000) The protein data bank. Nucleic Acids Res 28:235–242
Berendsen HJC, Van der Spoel D, Van Drunen R (1995) GROMACS: a message-passing parallel molecular dynamics implementation. Comput Phys Commun 91:43–56
Laskowski RA, MacArthur MW, Moss DS, Thornton JM (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr 26:283–291
Colovos C, Yeates TO (1993) Verification of protein structures: patterns of nonbonded atomic interactions. Protein Sci 2:1511–1519
Markus W, Manfred JS (2007) ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res 35:W407–W410
Krissinel E, Henrick H (2004) Secondary-structure matching (PDBeFold), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr D60:2256–2268
Ying CL, Chia CW, Ih SC, Jhao LJ, Jih HL, Chun WT (2013) TIPdb: a database of anticancer, antiplatelet, and antituberculosis phytochemicals from indigenous plants in Taiwan. Sci World J 2013:1–4
Sheeba Veluthoor et al. (2012) Phytochemicals: in pursuit of antitubercular drugs. In: Atta-ur-Rahman (Eds.). Studies in natural products chemistry. Elsevier Vol 38, pp. 417–463
Garcia A, Bocanegra GV, Palma-Nicolas JP, Rivera G (2012) Recent advances in antitubercular natural products. Eur J Med Chem 49:1–23
Kishore N, Mishra BB, Tripathi V, Tiwari VK (2009) Alkaloids as potential anti-tubercular agents. Fitoterapia 80:149–163
Gautam R, Saklani A, Jachak SM (2007) Indian medicinal plants as a source of antimycobacterial agents. J Ethnopharmacol 110:200–234
Bolton E, Wang Y et al. (2008) PubChem: integrated platform of small molecules and biological activities. Annual reports in computational chemistry, volume 4. American Chemical Society, Washington, DC
Jens S, Johann G, Gerhard K (1994) Comparison of automatic three-dimensional model builders using 639 X-ray structures. J Chem Inf Comput Sci 34:1000–1008
Thompson MA (2004) Molecular docking using arguslab: an efficient shape- based search algorithm and the AScore scoring function. Fall ACS Meeting, Philadelphia
Trott O, Olson AJ (2010) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading. J Comput Chem 31:455–461
Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ (2009) Autodock4 and AutoDockTools4: automated docking with selective receptor flexiblity. J Comp Chem 16:2785–2791
Stefano Forli Raccoon|AutoDock VS: an automated tool for preparing AutoDock virtual screenings. http://autodock.scripps.edu/resources/raccoon
Edward HK, Li D (2008) Drug-like properties: concepts, structure design and methods: from ADME to toxicity optimization. Academic, New York
Lipinski CA, Lombardo F, Dominy BW, Feeney PJ (2001) Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev 46:3–26
Filimonov D, Poroikov V (1996) Bioactive compound design: possibilities for industrial use. PASS: computerized prediction of biological activity spectra for chemical substances. BIOS Scientific, pp 44–56
Schüttelkopf AW, Van Aalten DMF (2004) PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr D60:1355–1363
Collins L, Franzblau SG (1997) Microplate alamar blue assay versus BACTEC 460 system for high- throughput screening of compounds against Mycobacterium tuberculosis and Mycobacterium avium. Antimicrob Agents Chemother 41(5):1004–1009
Lew JM, Kapopoulou A, Jones LM, Cole ST (2011) TubercuList, 10 years after. Tuberculosis (Edinb) 91(1):1–7
Takayama K, Kilburn JO (1989) Inhibition of synthesis of arabinogalactan by ethambutol in Mycobacterium smegmatis. Antimicrob Agents Chemother 33(9):1493–1499
Katrin P, Patric S, Kristina L, Per A (1997) Polar molecular surface properties predict the intestinal absorption of drugs in humans. Pharm Res 14:568–571
Joe D, Zheng O, Jeffery T, Andrew B, Yaron T, Jie L (2006) CASTp: computed atlas of surface topography of proteins with structural and topographical mapping of functionally annotated residues. Nucleic Acids Res 34:W116–W118
Bhargava S, Tyagi AK, Tyagi JS (1990) tRNA genes in mycobacteria: organization and molecular cloning. J Bacteriol 172:2930–2934
Collins DM, De Lisle GW (1984) DNA restriction endonuclease analysis of Mycobacterium tuberculosis and Mycobacterium bovis BCG. J Gen Microbiol 130:1019–1021
Imaeda T (1985) Deoxyribonucleic acid relatedness among strains of Mycobacterium tuberculosis, Mycobacterium bovis BCG, Mycobacterium microti and Mycobacterium africanum. Int J Syst Bacteriol 35:147–150
Acknowledgments
We acknowledge CDRI for assay service and VIT University for the computing facility. We thank all the anonymous reviewers for their valuable comments and suggestions.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
S. Fig. 1
Rhamnose biosynthesis pathway (DOCX 66 kb)
S. Fig. 2
2D structures of Phytocompounds used in the study (DOCX 434 kb)
S. Fig. 3
RmlC modeled using S. enterica template. (a) RmlC modeled (b) modeled RmlC superimposed with the crystal structure from M.tuberculosis (1UPI) (c) modeled RmlC superimposed with the crystal structure from S. enterica (1DZR). Cyan — modeled, purple — 1UPI, green — 1DZR (DOCX 315 kb)
S. Fig. 4
a: Backbone RMSD of the proteins and b: Potential energy plot of the proteins (brown — RmlA, violet — RmlB, cyan — RmlC, and orange — RmlD) (DOCX 99 kb)
S. Fig. 5
Multiple sequence alignment of RmlA sequences from Mycobacterium tuberculosis, Neisseria meningitides, Haemophilus parainfluenzae, Salmonella enterica, Klebsiella pneumoniae, Escherichia coli, Pseudomonas aeruginosa, and Streptococcus pneumoniae (DOCX 132 kb)
S. Fig. 6
Multiple sequence alignment of RmlB sequences from Mycobacterium tuberculosis, Neisseria meningitides, Haemophilus parainfluenzae, Salmonella enterica, Klebsiella pneumoniae, Escherichia coli, Pseudomonas aeruginosa, and Streptococcus pneumoniae (DOCX 269 kb)
S. Fig. 7
Multiple sequence alignment of RmlD sequences from Mycobacterium tuberculosis, Neisseria meningitides, Haemophilus parainfluenzae, Salmonella enterica, Klebsiella pneumoniae, Escherichia coli, Pseudomonas aeruginosa, and Streptococcus pneumonia (DOCX 221 kb)
S. Fig. 8
RMSD of the protein when bound to inhibitor and substrate. a: RmlA, b:RmlB, c: RmlC, and d: RmlD (DOCX 56 kb)
S. Fig. 9
RMSF of the binding pocket residues when bound to inhibitor and substrate. a: RmlA, b:RmlB, c: RmlC, and d: RmlD (DOCX 73 kb)
S. Fig. 10
Pair-wise alignment of RmlA sequences from H39Rv and H37Ra strains (DOCX 39 kb)
S. Fig. 11
Pair-wise alignment of RmlB sequences from H39Rv and H37Ra strains (DOCX 44 kb)
S. Fig. 12
Pair-wise alignment of RmlC sequences from H39Rv and H37Ra strains (DOCX 33 kb)
S. Fig. 13
Pair-wise alignment of RmlD sequences from H39Rv and H37Ra strains (DOCX 42 kb)
Rights and permissions
About this article
Cite this article
Sundarrajan, S., Lulu, S. & Arumugam, M. Computational evaluation of phytocompounds for combating drug resistant tuberculosis by multi-targeted therapy. J Mol Model 21, 247 (2015). https://doi.org/10.1007/s00894-015-2785-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00894-015-2785-z