Abstract
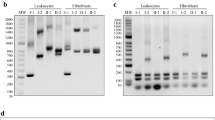
Friedreich ataxia (FRDA) is an autosomal recessive neurodegenerative disease most commonly caused by a GAA trinucleotide repeat expansion in the first intron of FXN, which reduces expression of the mitochondrial protein frataxin. Approximately 98% of individuals with FRDA are homozygous for GAA expansions, with the remaining 2% compound heterozygotes for a GAA expansion and a point mutation within FXN. Two siblings with early onset of symptoms experienced rapid loss of ambulation by 8 and 10 years. Diagnostic testing for FRDA demonstrated one GAA repeat expansion of 1010 repeats and one non-expanded allele. Sequencing all five exons of FXN identified a novel deletion-insertion mutation in exon 3 (c.371_376del6ins15), which results in a modified frataxin protein sequence at amino acid positions 124–127. Specifically, the amino acid sequence changes from DVSF to VHLEDT, increasing frataxin from 211 residues to 214. Using the known structure of human frataxin, a theoretical 3D model of the mutant protein was developed. In the event that the modified protein is expressed and stable, it is predicted that the acidic interface of frataxin, known to be involved in iron binding and interactions with the iron–sulphur cluster assembly factor IscU, would be impaired.



Similar content being viewed by others
References
Pandolfo M (2006) In: Wells RD, Ashizawa T (eds) Friedreich's ataxia. in Genetic Instabilities and Neurological Diseases. Academic, New York, pp 277–296
Delatycki MB, Williamson R, Forrest SM (2000) Friedreich ataxia: an overview. J Med Genet 37:1–8
Puccio H, Koenig M (2002) Friedreich ataxia: a paradigm for mitochondrial diseases. Curr Opin Genet Dev 12:272–277
Wilson CL, Fahey MC, Corben LA et al (2007) Quality of life in Friedreich ataxia: what clinical, social and demographic factors are important? Eur J Neurol 14:1040–1047
Delatycki MB, Paris DB, Gardner RJ et al (1999) Clinical and genetic study of Friedreich ataxia in an Australian population. Amer J of Med Genet 87:168–174
Cruz-Marino T, Gonzalez-Zaldivar Y, Laffita-Mesa JM et al (2010) Uncommon features in Cuban families affected with Friedreich ataxia. Neurosci Lett 472:85–89
Harding AE (1981) Friedreich's ataxia: a clinical and genetic study of 90 families with an analysis of early diagnostic criteria and intrafamilial clustering of clinical features. Brain 104:589–620
Porter N, Downes SM, Fratter C, Anslow P, Nemeth AH (2007) Catastrophic visual loss in a patient with Friedreich ataxia. Arch Ophthalmol 125:273–274
Fahey MC, Cremer PD, Aw ST et al (2008) Vestibular, saccadic and fixation abnormalities in genetically confirmed Friedreich ataxia. Brain 131:1035–1045
Rance G, Corben L, Barker E et al (2010) Auditory perception in individuals with Friedreich's ataxia. Audiol Neurootol 15:229–240
Rance G, Fava R, Baldock H et al (2008) Speech perception ability in individuals with Friedreich ataxia. Brain 131:2002–2012
Corben LA, Delatycki MB, Bradshaw JL et al (2010) Impairment in motor reprogramming in Friedreich ataxia reflecting possible cerebellar dysfunction. J Neurol 257:782–791
Fielding J, Corben L, Cremer P, Millist L, White O, Delatycki M (2010) Disruption to higher order processes in Friedreich ataxia. Neuropsychologia 48:235–242
Connelly T, Farmer JM, Lynch DR, Doty RL (2003) Olfactory dysfunction in degenerative ataxias. J Neurol Neurosurg & Psych 74:1435–1437
Lynch DR, Farmer JM, Balcer LJ, Wilson RB (2002) Friedreich ataxia: effects of genetic understanding on clinical evaluation and therapy.[see comment]. Arch Neurol 59:743–747
Rouault TA, Tong WH (2008) Iron-sulphur cluster biogenesis and human disease. Trends Genet 24:398–407
Santos R, Lefevre S, Sliwa D, Seguin A, Camadro JM, Lesuisse E (2010) Friedreich ataxia: molecular mechanisms, redox considerations, and therapeutic opportunities. Antioxid Redox Signal 13:651–690
Huang ML, Becker EM, Whitnall M, Rahmanto YS, Ponka P, Richardson DR (2009) Elucidation of the mechanism of mitochondrial iron loading in Friedreich's ataxia by analysis of a mouse mutant. Proc Natl Acad Sci U S A 106:16381–16386
De Biase I, Rasmussen A, Endres D et al (2007) Progressive GAA expansions in dorsal root ganglia of Friedreich's ataxia patients. Ann Neurol 61:55–60
Puccio H (2007) Conditional mouse models for Friedreich ataxia, a neurodegenerative disorder associating cardiomyopathy. Handb Exp Pharmacol 178:365–375
Sparaco M, Gaeta LM, Santorelli FM et al (2009) Friedreich's ataxia: oxidative stress and cytoskeletal abnormalities. J Neurol Sci 287:111–118
Campuzano V, Montermini L, Molto M et al (1996) Friedreich's ataxia: autosomal recessive disease caused by an intronic GAA triplet repeat expansion. Science 271:1423–1427
Gordon N (2000) Friedreich's ataxia and iron metabolism. Brain & Dev 22:465–468
Cossee M, Durr A, Schmitt M et al (1999) Friedreich's ataxia: point mutations and clinical presentation of compound heterozygotes. Ann Neurol 45:200–206
De Castro M, Garcia-Planells J, Monros E et al (2000) Genotype and phenotype analysis of Friedreich's ataxia compound heterozygous patients. Hum Genet 106:86–92
Delatycki MB, Knight M, Koenig M, Cossee M, Williamson R, Forrest SM (1999) G130V, a common FRDA point mutation, appears to have arisen from a common founder. Hum Genet 105:343–346
Subramony SH, May W, Lynch D et al (2005) Measuring Friedreich ataxia: Interrater reliability of a neurologic rating scale. Neurology 64:1261–1262
Soding J, Biegert A, Lupas AN (2005) The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res 33:W244–W248
Hildebrand A, Remmert M, Biegert A, Soding J (2009) Fast and accurate automatic structure prediction with HHpred. Proteins 77(Suppl 9):128–132
Dhe-Paganon S, Shigeta R, Chi YI, Ristow M, Shoelson SE (2000) Crystal structure of human frataxin. J Biol Chem 275:30753–30756
Berman HM, Westbrook J, Feng Z et al (2000) The Protein Data Bank. Nucleic Acids Res 28:235–242
Goodkin DE, Hertsgaard D, Seminary J (1988) Upper extremity function in multiple sclerosis: improving assessment sensitivity with box-and-block and nine-hole peg tests. Arch Phys Med Rehabil 69:850–854
Corben LA, Tai G, Wilson C, Collins V, Churchyard AJ, Delatycki MB (2010) A comparison of three measures of upper limb function in Friedreich ataxia. J Neurol 257:518–523
Bencze KZ, Kondapalli KC, Cook JD et al (2006) The structure and function of frataxin. Crit Rev Biochem Mol Biol 41:269–291
Bencze KZ, Yoon T, Millan-Pacheco C et al (2007) Human frataxin: iron and ferrochelatase binding surface. Chem Commun (Camb) 18:1798–1800
Stemmler TL, Lesuisse E, Pain D, Dancis A (2010) Frataxin and mitochondrial FeS cluster biogenesis. J Biol Chem 285:26737–26743
La Pean A, Jeffries N, Grow C, Ravina B, Di Prospero NA (2008) Predictors of progression in patients with Friedreich ataxia. Mov Disord 23:2026–2032
Streisinger G, Okada Y, Emrich J et al (1966) Frameshift mutations and the genetic code. This paper is dedicated to Professor Theodosius Dobzhansky on the occasion of his 66th birthday. Cold Spring Harb Symp Quant Biol 31:77–84
van Noort V, Worning P, Ussery DW, Rosche WA, Sinden RR (2003) Strand misalignments lead to quasipalindrome correction. Trends Genet 19:365–369
Rice P, Longden I, Bleasby A (2000) EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet 16:276–277
Correia AR, Wang T, Craig EA, Gomes CM (2010) Iron-binding activity in yeast frataxin entails a trade off with stability in the alpha1/beta1 acidic ridge region. Biochem J 426:197–203
Acknowledgements
The authors would like to sincerely thank the participating family as well as Gabrielle Wilson and Paul Lockhart for their assistance. This study was supported by funding from the Friedreich Ataxia Research Alliance, USA, the Friedreich Ataxia Research Association, Australasia, the Australian Rotary Health Research Fund, the North Brighton Rotary Club, the Collier Charitable Fund of Australia and the Victorian Government Operational Infrastructure Support Program. MBD is a National Health and Medical Research Council of Australia Practitioner Fellow.
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary materials
Below is the link to the electronic supplementary material.
Supplementary Fig. 1
Multiple amino acid sequence alignment of frataxin from various species. The alignment was performed using ClustalW2 on the Protein Information Resource (PRI) website (http://pir.georgetown.edu/pirwww/index.shtml). Amino acid residues known to bind iron are coloured red while residues involved in binding to IscU are blue and those that bind to both iron and IscU are green [36]. The site of the deletion–insertion mutation is in bold. (JPEG 213 kb)
Supplementary Fig. 2
Amino acid sequence alignment of the yeast and human Fe-S cluster scaffold proteins Isu1p and IscU. The alignment was performed using Clustal W2 on the Protein Information Resource (PRI) website (http://pir.georgetown.edu/pirwww/index.shtml). (JPEG 37 kb)
Rights and permissions
About this article
Cite this article
Evans-Galea, M.V., Corben, L.A., Hasell, J. et al. A novel deletion–insertion mutation identified in exon 3 of FXN in two siblings with a severe Friedreich ataxia phenotype. Neurogenetics 12, 307–313 (2011). https://doi.org/10.1007/s10048-011-0296-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10048-011-0296-3