Abstract
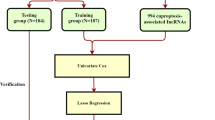
Cuproptosis has been recently used to indicate unique biological processes triggered by Cu action as a new term. This study aimed to explore the relationship between cuproptosis-related lncRNA and hepatocellular carcinoma (HCC) with regard to immunity and prognosis. RNA sequencing and the clinical data were downloaded from the TCGA database. The cuproptosis-related genes were sorted out through literature study. The cuproptosis-related IncRNA signature was identified by Cox regression analysis and the least absolute shrinkage and selection operator analysis. The K-M survival analysis, receiver operating characteristic analysis, and C-index analysis were adopted to evaluate the prognostic prediction performance of the signature. The functional enrichment, immune infiltration and tumor mutation analysis were further analyzed. Subsequently, we predicted the differences in chemosensitivity from tumor gene expression levels for some chemotherapy drugs. The prognostic signature consisting of 5 overall survival-related CUPlncRNAs. It showed an extraordinary ability to predict the prognoses of patients with HCC. The signature can predict the abundance of immune cell infiltration, immune functions, expression of immune checkpoint inhibitors, m6A genes, which was supported by the GO biological process and KEGG analysis. And it may also have a guiding effect in the sensitivity of different chemotherapeutic drugs and tumor mutation burden. We constructed a new cuproptosis-related lncRNA signature for HCC patients. The model can be used for prognostic prediction and immune evaluation, providing a reference for immunotherapies and targeted therapies.










Similar content being viewed by others
Data availability
Publicly available datasets were analyzed in this study. All datasets presented in this.
References
Sung H, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–49. https://doi.org/10.3322/caac.21660.
Petrick JL, et al. International trends in hepatocellular carcinoma incidence, 1978–2012. Int J Cancer. 2020;147:317–30. https://doi.org/10.1002/ijc.32723.
Mittal S, El-Serag H. Epidemiology of hepatocellular carcinoma: consider the population. J Clin Gastroenterol. 2013. https://doi.org/10.1097/MCG.0b013e3182872f29.
Llovet JM, et al. Hepatocellular carcinoma. Nat Rev Dis Primers. 2016;2:16018. https://doi.org/10.1038/nrdp.2016.18.
Petrick J, et al. International trends in hepatocellular carcinoma incidence, 1978–2012. Int J Cancer. 2020;147:317–30. https://doi.org/10.1002/ijc.32723.
Cucarull B, et al. Hepatocellular carcinoma: molecular pathogenesis and therapeutic advances. Cancers. 2022. https://doi.org/10.3390/cancers14030621.
Oliveri V. Selective targeting of cancer cells by copper ionophores: an overview. Front Mol Biosci. 2022;9:841814. https://doi.org/10.3389/fmolb.2022.841814.
Ruiz L, Libedinsky A, Elorza A. Role of copper on mitochondrial function and metabolism. Front Mol Biosci. 2021;8:711227. https://doi.org/10.3389/fmolb.2021.711227.
Basu S, et al. Heavy and trace metals in carcinoma of the gallbladder. World J Surg. 2013;37:2641–6. https://doi.org/10.1007/s00268-013-2164-9.
Ding X, et al. Analysis of serum levels of 15 trace elements in breast cancer patients in Shandong, China. Environ Sci Pollut Res Int. 2015;22:7930–5. https://doi.org/10.1007/s11356-014-3970-9.
Pavithra V, et al. Serum levels of metal ions in female patients with breast cancer. J Clin Diagn Res JCDR. 2015;9:BC25-c27. https://doi.org/10.7860/jcdr/2015/11627.5476.
Baltaci A, Dundar T, Aksoy F, Mogulkoc R. Changes in the serum levels of trace elements before and after the operation in thyroid cancer patients. Biol Trace Elem Res. 2017;175:57–64. https://doi.org/10.1007/s12011-016-0768-2.
Stepien M, et al. Pre-diagnostic copper and zinc biomarkers and colorectal cancer risk in the European prospective investigation into cancer and nutrition cohort. Carcinogenesis. 2017;38:699–707. https://doi.org/10.1093/carcin/bgx051.
Zhang X, Yang Q. Association between serum copper levels and lung cancer risk: a meta-analysis. J Int Med Res. 2018;46:4863–73. https://doi.org/10.1177/0300060518798507.
Chen F, et al. Serum copper and zinc levels and the risk of oral cancer: a new insight based on large-scale case-control study. Oral Dis. 2019;25:80–6. https://doi.org/10.1111/odi.12957.
Aubert L, et al. Copper bioavailability is a KRAS-specific vulnerability in colorectal cancer. Nat Commun. 2020;11:3701. https://doi.org/10.1038/s41467-020-17549-y.
Maslah H, Skarbek C, Pethe S, Labruère R. Anticancer boron-containing prodrugs responsive to oxidative stress from the tumor microenvironment. Eur J Med Chem. 2020;207:112670. https://doi.org/10.1016/j.ejmech.2020.112670.
Michniewicz F, et al. Copper: an intracellular achilles’ heel allowing the targeting of epigenetics, kinase pathways, and cell metabolism in cancer therapeutics. ChemMedChem. 2021;16:2315–29. https://doi.org/10.1002/cmdc.202100172.
Steinbrueck A, et al. Transition metal chelators, pro-chelators, and ionophores as small molecule cancer chemotherapeutic agents. Chem Soc Rev. 2020;49:3726–47. https://doi.org/10.1039/c9cs00373h.
Hunsaker E, Franz K. Emerging opportunities to manipulate metal trafficking for therapeutic benefit. Inorg Chem. 2019;58:13528–45. https://doi.org/10.1021/acs.inorgchem.9b01029.
Lelièvre P, Sancey L, Coll JL, Deniaud A, Busser B. The multifaceted roles of copper in cancer: a trace metal element with dysregulated metabolism, but also a target or a bullet for therapy. Cancers. 2020. https://doi.org/10.3390/cancers12123594.
Li Y. Copper homeostasis: emerging target for cancer treatment. IUBMB Life. 2020;72:1900–8. https://doi.org/10.1002/iub.2341.
Ge E, et al. Connecting copper and cancer: from transition metal signalling to metalloplasia. Nat Rev Cancer. 2022;22:102–13. https://doi.org/10.1038/s41568-021-00417-2.
Kung JT, Colognori D, Lee JT. Long noncoding RNAs: past, present, and future. Genetics. 2013;193:651–69. https://doi.org/10.1534/genetics.112.146704.
Li Y, et al. Identification of cancer risk lncRNAs and cancer risk pathways regulated by cancer risk lncRNAs based on genome sequencing data in human cancers. Sci Rep. 2016;6:39294. https://doi.org/10.1038/srep39294.
Castro-Oropeza R, Melendez-Zajgla J, Maldonado V, Vazquez-Santillan K. The emerging role of lncRNAs in the regulation of cancer stem cells. Cell Oncol (Dordr). 2018;41:585–603. https://doi.org/10.1007/s13402-018-0406-4.
Jiang M, Ni J, Cui W, Wang B, Zhuo W. Emerging roles of lncRNA in cancer and therapeutic opportunities. Am J Cancer Res. 2019;9:1354–66.
Hu Q, et al. Oncogenic lncRNA downregulates cancer cell antigen presentation and intrinsic tumor suppression. Nat Immunol. 2019;20:835–51. https://doi.org/10.1038/s41590-019-0400-7.
Xu M, et al. LncRNA SATB2-AS1 inhibits tumor metastasis and affects the tumor immune cell microenvironment in colorectal cancer by regulating SATB2. Mol Cancer. 2019;18:135. https://doi.org/10.1186/s12943-019-1063-6.
Forner A, Reig M, Bruix J. Hepatocellular carcinoma. Lancet (London, England). 2018;391:1301–14. https://doi.org/10.1016/s0140-6736(18)30010-2.
Gao W, Chen X, Chi W, Xue M. Long non-coding RNA MKLN1-AS aggravates hepatocellular carcinoma progression by functioning as a molecular sponge for miR-654-3p, thereby promoting hepatoma-derived growth factor expression. Int J Mol Med. 2020;46:1743–54. https://doi.org/10.3892/ijmm.2020.4722.
Pan G, et al. ETS Proto-Oncogene 1-activated muskelin 1 antisense RNA drives the malignant progression of hepatocellular carcinoma by targeting miR-22-3p to upregulate ETS Proto-Oncogene 1. Bioengineered. 2022;13:1346–58. https://doi.org/10.1080/21655979.2021.2017565.
Tang P, et al. Identifying a hypoxia-related long non-coding RNAs signature to improve the prediction of prognosis and immunotherapy response in hepatocellular carcinoma. Front Genet. 2021;12:785185. https://doi.org/10.3389/fgene.2021.785185.
Deng X, et al. Identification of a five-autophagy-related-lncRNA signature as a novel prognostic biomarker for hepatocellular carcinoma. Front Mol Biosci. 2020;7:611626. https://doi.org/10.3389/fmolb.2020.611626.
Wu Z, Li Z, Yang D, Liu J. Development and validation of a pyroptosis-related long non-coding RNA signature for hepatocellular carcinoma. Front Cell Dev Biol. 2021;9:713925. https://doi.org/10.3389/fcell.2021.713925.
Fang C, et al. Ferroptosis-related lncRNA signature predicts the prognosis and immune microenvironment of hepatocellular carcinoma. Sci Rep. 2022;12:6642. https://doi.org/10.1038/s41598-022-10508-1.
Zhang Z, et al. Construction and validation of a ferroptosis-related lncRNA signature as a novel biomarker for prognosis, immunotherapy and targeted therapy in hepatocellular carcinoma. Front Cell Dev Biol. 2022;10:792676. https://doi.org/10.3389/fcell.2022.792676.
Wei J, Zeng Y, Gao X, Liu T. A novel ferroptosis-related lncRNA signature for prognosis prediction in gastric cancer. BMC Cancer. 2021;21:1221. https://doi.org/10.1186/s12885-021-08975-2.
Ma W, et al. Immune-related lncRNAs as predictors of survival in breast cancer: a prognostic signature. J Transl Med. 2020;18:442. https://doi.org/10.1186/s12967-020-02522-6.
Han T, et al. Identification of a robust signature for clinical outcomes and immunotherapy response in gastric cancer: based on N6-methyladenosine related long noncoding RNAs. Cancer Cell Int. 2021;21:432. https://doi.org/10.1186/s12935-021-02146-w.
Wang Y, et al. Comprehensive analysis of tumor immune microenvironment and prognosis of m6A-related lncRNAs in gastric cancer. BMC Cancer. 2022;22:316. https://doi.org/10.1186/s12885-022-09377-8.
Chen W, et al. Identification of ferroptosis-related long noncoding RNA and construction of a novel prognostic signature for gastric cancer. Dis Markers. 2021;2021:7724997. https://doi.org/10.1155/2021/7724997.
Huang J, Chen W, Chen C, Jie Z, Xiao T. Comprehensive analysis and prognosis prediction of n6-methyladenosine-related lncRNAs in immune microenvironment infiltration of gastric cancer. Int J Gen Med. 2022;15:2629–43. https://doi.org/10.2147/ijgm.S349399.
Yu Z, Zhu Z. N6-Methyladenosine related long non-coding RNAs and immune cell infiltration in the tumor microenvironment of gastric cancer. Biol Proced Online. 2021;23:15. https://doi.org/10.1186/s12575-021-00152-w.
Li H, et al. A Novel ferroptosis-related gene signature predicts overall survival of breast cancer patients. Biology. 2021. https://doi.org/10.3390/biology10020151.
Zhu G, et al. Prognostic value of ferroptosis-related genes in patients with lung adenocarcinoma. Thoracic cancer. 2021;12:1890–9. https://doi.org/10.1111/1759-7714.13998.
Zhu L, et al. Identification the ferroptosis-related gene signature in patients with esophageal adenocarcinoma. Cancer Cell Int. 2021;21:124. https://doi.org/10.1186/s12935-021-01821-2.
Liu J, et al. Construction and external validation of a ferroptosis-related gene signature of predictive value for the overall survival in bladder cancer. Front Mol Biosci. 2021;8:675651. https://doi.org/10.3389/fmolb.2021.675651.
Crow J, Samuel G, Godwin A. Beyond tumor mutational burden: potential and limitations in using exosomes to predict response to immunotherapy. Expert Rev Mol Diagn. 2019;19:1079–88. https://doi.org/10.1080/14737159.2020.1688144.
Yarchoan M, Hopkins A, Jaffee E. Tumor mutational burden and response rate to PD-1 inhibition. N Engl J Med. 2017;377:2500–1. https://doi.org/10.1056/NEJMc1713444.
Chan T, et al. Development of tumor mutation burden as an immunotherapy biomarker: utility for the oncology clinic. Ann Oncol Off J Eur Soc Med Oncol. 2019;30:44–56. https://doi.org/10.1093/annonc/mdy495.
Zhu J, et al. Association between tumor mutation burden (TMB) and outcomes of cancer patients treated with PD-1/PD-L1 inhibitions: a meta-analysis. Front Pharmacol. 2019;10:673. https://doi.org/10.3389/fphar.2019.00673.
Jang B, Han W, Kim I. Tumor mutation burden, immune checkpoint crosstalk and radiosensitivity in single-cell RNA sequencing data of breast cancer. Radiother Oncol J Eur Soc Ther Radiol Oncol. 2020;142:202–9. https://doi.org/10.1016/j.radonc.2019.11.003.
Samstein R, et al. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat Genet. 2019;51:202–6. https://doi.org/10.1038/s41588-018-0312-8.
Yin L, Zhou L, Xu R. Identification of tumor mutation burden and immune infiltrates in hepatocellular carcinoma based on multi-omics analysis. Front Mol Biosci. 2020;7:599142. https://doi.org/10.3389/fmolb.2020.599142.
Gao X, et al. Genetic expression and mutational profile analysis in different pathologic stages of hepatocellular carcinoma patients. BMC Cancer. 2021;21:786. https://doi.org/10.1186/s12885-021-08442-y.
Kancherla V, et al. TP53Genomic analysis revealed new oncogenic signatures in -mutant hepatocellular carcinoma. Front Genet. 2018;9:2. https://doi.org/10.3389/fgene.2018.00002.
Oh J, et al. TTNSpontaneous mutations in the single gene represent high tumor mutation burden. NPJ Genom Med. 2020;5:33. https://doi.org/10.1038/s41525-019-0107-6.
Wang Z, Wang C, Lin S, Yu X. Effect of TTN mutations on immune microenvironment and efficacy of immunotherapy in lung adenocarcinoma patients. Front Oncol. 2021;11:725292. https://doi.org/10.3389/fonc.2021.725292.
Zou S, Ye J, Hu S, Wei Y, Xu J. TTNMutations in the gene are a prognostic factor for patients with lung squamous Cell carcinomas. Int J Gen Med. 2022;15:19–31. https://doi.org/10.2147/ijgm.S343259.
Xu L, et al. Genomic and transcriptional heterogeneity of multifocal hepatocellular carcinoma. Ann Oncol Off J Eur Soc Med Oncol. 2019;30:990–7. https://doi.org/10.1093/annonc/mdz103.
Dragani T. Risk of HCC: genetic heterogeneity and complex genetics. J Hepatol. 2010;52:252–7. https://doi.org/10.1016/j.jhep.2009.11.015.
Zhang Q, et al. Circulating tumor cells in hepatocellular carcinoma: single-cell based analysis, preclinical models, and clinical applications. Theranostics. 2020;10:12060–71. https://doi.org/10.7150/thno.48918.
Jeng K, Chang C, Jeng W, Sheen I, Jeng C. Heterogeneity of hepatocellular carcinoma contributes to cancer progression. Crit Rev Oncol Hematol. 2015;94:337–47. https://doi.org/10.1016/j.critrevonc.2015.01.009.
Acknowledgements
The authors gratefully acknowledge contributions from the TCGA network and the reviewers for their helpful comments on this study.
Funding
This study was supported by National Science and Technology Major Project of China (2017ZX10203201002-002).
Author information
Authors and Affiliations
Contributions
GJH and ZQY contributed to study conception and design and drafting of manuscript; ZQY and HY were involved in acquisition of data; ZQY, HY, XY and LYM contributed to analysis and interpretation of data; ZQY, HY, XY, LYM and GJH contributed to the writing, review, revision of the manuscript. All authors contributed to the article and approved the submitted version.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Consent for publication
The work has not been published previously, and it is not under consideration for publication elsewhere.
Ethical approval
We performed this research though publicly available datasets. There was no ethical approval was required.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhang, Q., Huang, Y., Xia, Y. et al. Cuproptosis-related lncRNAs predict the prognosis and immune response in hepatocellular carcinoma. Clin Exp Med 23, 2051–2064 (2023). https://doi.org/10.1007/s10238-022-00892-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10238-022-00892-3