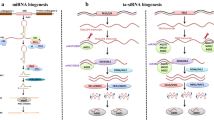
Abstract
Small RNA-mediated gene silencing encompasses diverse developmental events, stress responses, defense against pathogens, and maintenance of genome integrity. Extensive studies in model organisms have unveiled the molecular mechanisms underpinning the RNA silencing phenomena, and the accumulating knowledge have characterized the intricate pathways and the repertoire of proteins responsible for the actions of small RNAs characterized as microRNAs (miRNAs) or small interfering RNAs (siRNAs). Although the single-stranded, matured guide small RNAs direct the effector ribonucleoprotein complexes to induce gene silencing in sequence-specific manner, the double-stranded intermediate, the small RNA duplexes, which are processed as nascent products of the RNase III enzyme activities, act as key to determine the downstream molecular pathways and the fate of small RNAs. Based at the small RNA duplex-centered view, this review describes the recent advances in understanding the small RNA pathways in plants.


Similar content being viewed by others
References
Adenot X, Elmayan T, Lauressergues D, Boutet S, Bouche N, Gasciolli V, Vaucheret H (2006) DRB4-dependent TAS3 trans-acting siRNAs control leaf morphology through AGO7. Curr Biol 16:927–932
Arribas-Hernandez L, Kielpinski LJ, Brodersen P (2016a) mRNA Decay of Most Arabidopsis miRNA Targets Requires Slicer Activity of AGO1. Plant Physiol 171:2620–2632
Arribas-Hernandez L, Marchais A, Poulsen C, Haase B, Hauptmann J, Benes V, Meister G, Brodersen P (2016b) The Slicer Activity of ARGONAUTE1 Is Required Specifically for the Phasing, Not Production, of Trans-Acting Short Interfering RNAs in Arabidopsis. Plant Cell 28:1563–1580
Baumberger N, Baulcombe DC (2005) Arabidopsis ARGONAUTE1 is an RNA Slicer that selectively recruits microRNAs and short interfering RNAs. Proc Natl Acad Sci USA 102:11928–11933
Berardini TZ, Bollman K, Sun H, Poethig RS (2001) Regulation of vegetative phase change in Arabidopsis thaliana by cyclophilin 40. Science 291:2405–2407
Bohmert K, Camus I, Bellini C, Bouchez D, Caboche M, Benning C (1998) AGO1 defines a novel locus of Arabidopsis controlling leaf development. EMBO J 17:170–180
Bologna NG, Voinnet O (2014) The diversity, biogenesis, and activities of endogenous silencing small RNAs in Arabidopsis. Annu Rev Plant Biol 65:473–503
Bologna NG, Mateos JL, Bresso EG, Palatnik JF (2009) A loop-to-base processing mechanism underlies the biogenesis of plant microRNAs miR319 and miR159. EMBO J 28:3646–3656
Bologna NG, Schapire AL, Zhai J, Chorostecki U, Boisbouvier J, Meyers BC, Palatnik JF (2013) Multiple RNA recognition patterns during microRNA biogenesis in plants. Genome Res 23:1675–1689
Carbonell A, Carrington JC (2015) Antiviral roles of plant ARGONAUTES. Curr Opin Plant Biol 27:111–117
Carbonell A, Fahlgren N, Garcia-Ruiz H, Gilbert KB, Montgomery TA, Nguyen T, Cuperus JT, Carrington JC (2012) Functional analysis of three Arabidopsis ARGONAUTES using slicer-defective mutants. Plant Cell 24:3613–3629
Chen HM, Chen LT, Patel K, Li YH, Baulcombe DC, Wu SH (2010) 22-Nucleotide RNAs trigger secondary siRNA biogenesis in plants. Proc Natl Acad Sci USA 107:15269–15274
Creasey KM, Zhai J, Borges F, Van Ex F, Regulski M, Meyers BC, Martienssen RA (2014) miRNAs trigger widespread epigenetically activated siRNAs from transposons in Arabidopsis. Nature 508:411–415
Csorba T, Kontra L, Burgyan J (2015) viral silencing suppressors: tools forged to fine-tune host-pathogen coexistence. Virology 479–480:85–103
Cuperus JT, Carbonell A, Fahlgren N, Garcia-Ruiz H, Burke RT, Takeda A, Sullivan CM, Gilbert SD, Montgomery TA, Carrington JC (2010) Unique functionality of 22-nt miRNAs in triggering RDR6-dependent siRNA biogenesis from target transcripts in Arabidopsis. Nat Struct Mol Biol 17:997–1003
Deleris A, Gallego-Bartolome J, Bao J, Kasschau KD, Carrington JC, Voinnet O (2006) Hierarchical action and inhibition of plant Dicer-like proteins in antiviral defense. Science 313:68–71
Dong Z, Han MH, Fedoroff N (2008) The RNA-binding proteins HYL1 and SE promote accurate in vitro processing of pri-miRNA by DCL1. Proc Natl Acad Sci USA 105:9970–9975
Eamens AL, Smith NA, Curtin SJ, Wang MB, Waterhouse PM (2009) The Arabidopsis thaliana double-stranded RNA binding protein DRB1 directs guide strand selection from microRNA duplexes. RNA 15:2219–2235
Earley KW, Poethig RS (2011) Binding of the cyclophilin 40 ortholog SQUINT to Hsp90 protein is required for SQUINT function in Arabidopsis. J Biol Chem 286:38184–38189
Elkayam E, Kuhn CD, Tocilj A, Haase AD, Greene EM, Hannon GJ, Joshua-Tor L (2012) The structure of human argonaute-2 in complex with miR-20a. Cell 150:100–110
Endo Y, Iwakawa HO, Tomari Y (2013) Arabidopsis ARGONAUTE7 selects miR390 through multiple checkpoints during RISC assembly. EMBO Rep 14:652–658
Fahlgren N, Montgomery TA, Howell MD, Allen E, Dvorak SK, Alexander AL, Carrington JC (2006) Regulation of AUXIN RESPONSE FACTOR3 by TAS3 ta-siRNA affects developmental timing and patterning in Arabidopsis. Curr Biol 16:939–944
Fang X, Qi Y (2016) RNAi in Plants: an Argonaute-Centered View. Plant Cell 28:272–285
Frank F, Sonenberg N, Nagar B (2010) Structural basis for 5′-nucleotide base-specific recognition of guide RNA by human AGO2. Nature 465:818–822
Frank F, Hauver J, Sonenberg N, Nagar B (2012) Arabidopsis Argonaute MID domains use their nucleotide specificity loop to sort small RNAs. EMBO J 31:3588–3595
Garcia D, Collier SA, Byrne ME, Martienssen RA (2006) Specification of leaf polarity in Arabidopsis via the trans-acting siRNA pathway. Curr Biol 16:933–938
Ghildiyal M, Xu J, Seitz H, Weng Z, Zamore PD (2010) Sorting of Drosophila small silencing RNAs partitions microRNA* strands into the RNA interference pathway. RNA 16:43–56
Guy NC, Garcia YA, Sivils JC, Galigniana MD, Cox MB (2015) Functions of the Hsp90-binding FKBP immunophilins. Subcell Biochem 78:35–68
Ha M, Kim VN (2014) Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol 15:509–524
Huang Y, Ji L, Huang Q, Vassylyev DG, Chen X, Ma JB (2009) Structural insights into mechanisms of the small RNA methyltransferase HEN1. Nature 461:823–827
Hunter C, Willmann MR, Wu G, Yoshikawa M, de la Luz Gutierrez-Nava M, Poethig SR (2006) Trans-acting siRNA-mediated repression of ETTIN and ARF4 regulates heteroblasty in Arabidopsis. Development 133:2973–2981
Iki T, Yoshikawa M, Nishikiori M, Jaudal MC, Matsumoto-Yokoyama E, Mitsuhara I, Meshi T, Ishikawa M (2010) In vitro assembly of plant RNA-induced silencing complexes facilitated by molecular chaperone HSP90. Mol Cell 39:282–291
Iki T, Yoshikawa M, Meshi T, Ishikawa M (2012) Cyclophilin 40 facilitates HSP90-mediated RISC assembly in plants. EMBO J 31:267–278
Iwasaki S, Kobayashi M, Yoda M, Sakaguchi Y, Katsuma S, Suzuki T, Tomari Y (2010) Hsc70/Hsp90 chaperone machinery mediates ATP-dependent RISC loading of small RNA duplexes. Mol Cell 39:292–299
Iwasaki S, Sasaki HM, Sakaguchi Y, Suzuki T, Tadakuma H, Tomari Y (2015) Defining fundamental steps in the assembly of the Drosophila RNAi enzyme complex. Nature 521:533–536
Iwata Y, Takahashi M, Fedoroff NV, Hamdan SM (2013) Dissecting the interactions of SERRATE with RNA and DICER-LIKE 1 in Arabidopsis microRNA precursor processing. Nucleic Acids Res 41:9129–9140
Khvorova A, Reynolds A, Jayasena SD (2003) Functional siRNAs and miRNAs exhibit strand bias. Cell 115:209–216
Kidner CA, Martienssen RA (2004) Spatially restricted microRNA directs leaf polarity through ARGONAUTE1. Nature 428:81–84
Kobayashi H, Tomari Y (2016) RISC assembly: coordination between small RNAs and Argonaute proteins. Biochim Biophys Acta 1859:71–81
Kurihara Y, Takashi Y, Watanabe Y (2006) The interaction between DCL1 and HYL1 is important for efficient and precise processing of pri-miRNA in plant microRNA biogenesis. RNA 12:206–212
Lakatos L, Csorba T, Pantaleo V, Chapman EJ, Carrington JC, Liu YP, Dolja VV, Calvino LF, Lopez-Moya JJ, Burgyan J (2006) Small RNA binding is a common strategy to suppress RNA silencing by several viral suppressors. EMBO J 25:2768–2780
Lee WC, Lu SH, Lu MH, Yang CJ, Wu SH, Chen HM (2015) Asymmetric bulges and mismatches determine 20-nt microRNA formation in plants. RNA Biol 12:1054–1066
Leuschner PJ, Ameres SL, Kueng S, Martinez J (2006) Cleavage of the siRNA passenger strand during RISC assembly in human cells. EMBO Rep 7:314–320
Li J, Yang Z, Yu B, Liu J, Chen X (2005) Methylation protects miRNAs and siRNAs from a 3′-end uridylation activity in Arabidopsis. Curr Biol 15:1501–1507
Liu Q, Yao X, Pi L, Wang H, Cui X, Huang H (2009) The ARGONAUTE10 gene modulates shoot apical meristem maintenance and establishment of leaf polarity by repressing miR165/166 in Arabidopsis. Plant J 58:27–40
MacRae IJ, Zhou K, Doudna JA (2007) Structural determinants of RNA recognition and cleavage by Dicer. Nat Struct Mol Biol 14:934–940
Manavella PA, Hagmann J, Ott F, Laubinger S, Franz M, Macek B, Weigel D (2012a) Fast-forward genetics identifies plant CPL phosphatases as regulators of miRNA processing factor HYL1. Cell 151:859–870
Manavella PA, Koenig D, Weigel D (2012b) Plant secondary siRNA production determined by microRNA-duplex structure. Proc Natl Acad Sci USA 109:2461–2466
Martinez NJ, Chang HM, Borrajo Jde R, Gregory RI (2013) The co-chaperones Fkbp4/5 control Argonaute2 expression and facilitate RISC assembly. RNA 19:1583–1593
Matranga C, Tomari Y, Shin C, Bartel DP, Zamore PD (2005) Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell 123:607–620
Maunoury N, Vaucheret H (2011) AGO1 and AGO2 act redundantly in miR408-mediated Plantacyanin regulation. PLoS One 6:e28729
Mi S, Cai T, Hu Y, Chen Y, Hodges E, Ni F, Wu L, Li S, Zhou H, Long C et al (2008) Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5′ terminal nucleotide. Cell 133:116–127
Miyoshi K, Tsukumo H, Nagami T, Siomi H, Siomi MC (2005) Slicer function of Drosophila Argonautes and its involvement in RISC formation. Genes Dev 19:2837–2848
Miyoshi T, Takeuchi A, Siomi H, Siomi MC (2010) A direct role for Hsp90 in pre-RISC formation in Drosophila. Nat Struct Mol Biol 17:1024–1026
Mlotshwa S, Pruss GJ, Peragine A, Endres MW, Li J, Chen X, Poethig RS, Bowman LH, Vance V (2008) DICER-LIKE2 plays a primary role in transitive silencing of transgenes in Arabidopsis. PLoS One 3:e1755
Montgomery TA, Howell MD, Cuperus JT, Li D, Hansen JE, Alexander AL, Chapman EJ, Fahlgren N, Allen E, Carrington JC (2008) Specificity of ARGONAUTE7-miR390 interaction and dual functionality in TAS3 trans-acting siRNA formation. Cell 133:128–141
Morel JB, Godon C, Mourrain P, Beclin C, Boutet S, Feuerbach F, Proux F, Vaucheret H (2002) Fertile hypomorphic ARGONAUTE (ago1) mutants impaired in post-transcriptional gene silencing and virus resistance. Plant Cell 14:629–639
Nagano H, Fukudome A, Hiraguri A, Moriyama H, Fukuhara T (2014) Distinct substrate specificities of Arabidopsis DCL3 and DCL4. Nucleic Acids Res 42:1845–1856
Nguyen TA, Jo MH, Choi YG, Park J, Kwon SC, Hohng S, Kim VN, Woo JS (2015) Functional Anatomy of the Human Microprocessor. Cell 161:1374–1387
Okamura K, Liu N, Lai EC (2009) Distinct mechanisms for microRNA strand selection by Drosophila Argonautes. Mol Cell 36:431–444
Parent JS, Bouteiller N, Elmayan T, Vaucheret H (2015) Respective contributions of Arabidopsis DCL2 and DCL4 to RNA silencing. Plant J 81:223–232
Park JH, Shin C (2015) Slicer-independent mechanism drives small-RNA strand separation during human RISC assembly. Nucleic Acids Res 43:9418–9433
Park JE, Heo I, Tian Y, Simanshu DK, Chang H, Jee D, Patel DJ, Kim VN (2011) Dicer recognizes the 5′ end of RNA for efficient and accurate processing. Nature 475:201–205
Pumplin N, Voinnet O (2013) RNA silencing suppression by plant pathogens: defence, counter-defence and counter-counter-defence. Nat Rev Microbiol 11:745–760
Qi Y, Denli AM, Hannon GJ (2005) Biochemical specialization within Arabidopsis RNA silencing pathways. Mol Cell 19:421–428
Rand TA, Petersen S, Du F, Wang X (2005) Argonaute2 cleaves the anti-guide strand of siRNA during RISC activation. Cell 123:621–629
Ren G, Chen X, Yu B (2012a) Uridylation of miRNAs by hen1 suppressor1 in Arabidopsis. Curr Biol 22:695–700
Ren G, Xie M, Dou Y, Zhang S, Zhang C, Yu B (2012b) Regulation of miRNA abundance by RNA binding protein TOUGH in Arabidopsis. Proc Natl Acad Sci USA 109:12817–12821
Schirle NT, MacRae IJ (2012) The crystal structure of human Argonaute2. Science 336:1037–1040
Schott G, Mari-Ordonez A, Himber C, Alioua A, Voinnet O, Dunoyer P (2012) Differential effects of viral silencing suppressors on siRNA and miRNA loading support the existence of two distinct cellular pools of ARGONAUTE1. EMBO J 31:2553–2565
Schuck J, Gursinsky T, Pantaleo V, Burgyan J, Behrens SE (2013) AGO/RISC-mediated antiviral RNA silencing in a plant in vitro system. Nucleic Acids Res 41:5090–5103
Schwarz DS, Hutvagner G, Du T, Xu Z, Aronin N, Zamore PD (2003) Asymmetry in the assembly of the RNAi enzyme complex. Cell 115:199–208
Smith MR, Willmann MR, Wu G, Berardini TZ, Moller B, Weijers D, Poethig RS (2009) Cyclophilin 40 is required for microRNA activity in Arabidopsis. Proc Natl Acad Sci USA 106:5424–5429
Sorin C, Bussell JD, Camus I, Ljung K, Kowalczyk M, Geiss G, McKhann H, Garcion C, Vaucheret H, Sandberg G et al (2005) Auxin and light control of adventitious rooting in Arabidopsis require ARGONAUTE1. Plant Cell 17:1343–1359
Starega-Roslan J, Krol J, Koscianska E, Kozlowski P, Szlachcic WJ, Sobczak K, Krzyzosiak WJ (2011) Structural basis of microRNA length variety. Nucleic Acids Res 39:257–268
Tomari Y, Matranga C, Haley B, Martinez N, Zamore PD (2004) A protein sensor for siRNA asymmetry. Science 306:1377–1380
Tomari Y, Du T, Zamore PD (2007) Sorting of Drosophila small silencing RNAs. Cell 130:299–308
Tu B, Liu L, Xu C, Zhai J, Li S, Lopez MA, Zhao Y, Yu Y, Ramachandran V, Ren G et al (2015) Distinct and cooperative activities of HESO1 and URT1 nucleotidyl transferases in microRNA turnover in Arabidopsis. PLoS Genet 11:e1005119
Vargason JM, Szittya G, Burgyan J, Hall TM (2003) Size selective recognition of siRNA by an RNA silencing suppressor. Cell 115:799–811
Vazquez F, Blevins T, Ailhas J, Boller T, Meins F Jr (2008) Evolution of Arabidopsis MIR genes generates novel microRNA classes. Nucleic Acids Res 36:6429–6438
Wang X, Zhang S, Dou Y, Zhang C, Chen X, Yu B, Ren G (2015) Synergistic and independent actions of multiple terminal nucleotidyl transferases in the 3′ tailing of small RNAs in Arabidopsis. PLoS Genet 11:e1005091
Wilson RC, Tambe A, Kidwell MA, Noland CL, Schneider CP, Doudna JA (2015) Dicer-TRBP complex formation ensures accurate mammalian microRNA biogenesis. Mol Cell 57:397–407
Woehrer SL, Aronica L, Suhren JH, Busch CJ, Noto T, Mochizuki K (2015) A Tetrahymena Hsp90 co-chaperone promotes siRNA loading by ATP-dependent and ATP-independent mechanisms. EMBO J 34:559–577
Wu L, Zhang Q, Zhou H, Ni F, Wu X, Qi Y (2009) Rice MicroRNA effector complexes and targets. Plant Cell 21:3421–3435
Wu L, Zhou H, Zhang Q, Zhang J, Ni F, Liu C, Qi Y (2010) DNA methylation mediated by a microRNA pathway. Mol Cell 38:465–475
Ye K, Malinina L, Patel DJ (2003) Recognition of small interfering RNA by a viral suppressor of RNA silencing. Nature 426:874–878
Ye R, Wang W, Iki T, Liu C, Wu Y, Ishikawa M, Zhou X, Qi Y (2012) Cytoplasmic assembly and selective nuclear import of Arabidopsis Argonaute4/siRNA complexes. Mol Cell 46:859–870
Yoshikawa M (2013) Biogenesis of trans-acting siRNAs, endogenous secondary siRNAs in plants. Genes Genet Syst 88:77–84
Yoshikawa M, Iki T, Tsutsui Y, Miyashita K, Poethig RS, Habu Y, Ishikawa M (2013) 3′ fragment of miR173-programmed RISC-cleaved RNA is protected from degradation in a complex with RISC and SGS3. Proc Natl Acad Sci USA 110:4117–4122
Yu B, Yang Z, Li J, Minakhina S, Yang M, Padgett RW, Steward R, Chen X (2005) Methylation as a crucial step in plant microRNA biogenesis. Science 307:932–935
Zhai J, Zhao Y, Simon SA, Huang S, Petsch K, Arikit S, Pillay M, Ji L, Xie M, Cao X et al (2013) Plant microRNAs display differential 3′ truncation and tailing modifications that are ARGONAUTE1 dependent and conserved across species. Plant Cell 25:2417–2428
Zhang H, Kolb FA, Jaskiewicz L, Westhof E, Filipowicz W (2004) Single processing center models for human Dicer and bacterial RNase III. Cell 118:57–68
Zhang X, Zhao H, Gao S, Wang WC, Katiyar-Agarwal S, Huang HD, Raikhel N, Jin H (2011) Arabidopsis Argonaute 2 regulates innate immunity via miRNA393(*)-mediated silencing of a Golgi-localized SNARE gene, MEMB12. Mol Cell 42:356–366
Zhang X, Niu D, Carbonell A, Wang A, Lee A, Tun V, Wang Z, Carrington JC, Chang CE, Jin H (2014) ARGONAUTE PIWI domain and microRNA duplex structure regulate small RNA sorting in Arabidopsis. Nat Commun 5:5468
Zhang Z, Liu X, Guo X, Wang XJ, Zhang X (2016) Arabidopsis AGO3 predominantly recruits 24-nt small RNAs to regulate epigenetic silencing. Nat Plants 2:16049
Zhao Y, Yu Y, Zhai J, Ramachandran V, Dinh TT, Meyers BC, Mo B, Chen X (2012) The Arabidopsis nucleotidyl transferase HESO1 uridylates unmethylated small RNAs to trigger their degradation. Curr Biol 22:689–694
Zhu H, Hu F, Wang R, Zhou X, Sze SH, Liou LW, Barefoot A, Dickman M, Zhang X (2011) Arabidopsis Argonaute10 specifically sequesters miR166/165 to regulate shoot apical meristem development. Cell 145:242–256
Zhu H, Zhou Y, Castillo-Gonzalez C, Lu A, Ge C, Zhao YT, Duan L, Li Z, Axtell MJ, Wang XJ et al (2013) Bidirectional processing of pri-miRNAs with branched terminal loops by Arabidopsis Dicer-like1. Nat Struct Mol Biol 20:1106–1115
Acknowledgements
I thank Dr. Misato Ohtani and Dr. Reina Komiya for giving me the opportunity to publish this review related to the JPR symposium held in 2015.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The author declares that he has no conflict of interest.
Rights and permissions
About this article
Cite this article
Iki, T. Messages on small RNA duplexes in plants. J Plant Res 130, 7–16 (2017). https://doi.org/10.1007/s10265-016-0876-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10265-016-0876-2