-
PDF
- Split View
-
Views
-
Cite
Cite
Simon Vainberg, Charles W Condee, Robert J Steffan, Large-scale production of bacterial consortia for remediation of chlorinated solvent-contaminated groundwater, Journal of Industrial Microbiology and Biotechnology, Volume 36, Issue 9, 1 September 2009, Pages 1189–1197, https://doi.org/10.1007/s10295-009-0600-5
Close - Share Icon Share
Abstract
Chlorinated solvents such as perchloroethylene (PCE) and trichloroethylene (TCE) continue to be significant groundwater contaminants throughout the USA. In many cases efficient bioremediation of aquifers contaminated with these chemicals requires the addition of exogenous microorganisms, specifically members of the genus Dehalococcoides (DHC). This process is referred to as bioaugmentation. In this study a fed-batch fermentation process was developed for producing large volumes (to 3,200 L) of DHC-containing consortia suitable for treating contaminated aquifers. Three consortia enriched from three different sites were grown anaerobically with sodium lactate as an electron donor and PCE or TCE as an electron acceptor. DHC titers in excess of 1011 DHC/L could be reproducibly obtained at all scales tested and with all three of the enrichment cultures. The mean specific DHC growth rate for culture SDC-9™ was 0.036 ± 0.005 (standard error, SE)/h with a calculated mean doubling time of 19.3 ± 2.7 (SE) h. Finished cultures could be concentrated approximately tenfold by membrane filtration and stored refrigerated (4°C) for more that 40 days without measurable loss of activity. Dehalogenation of PCE by the fermented cultures was affected by pH with no measurable activity at pH <5.0.
Introduction
Chlorinated ethenes have been used extensively as industrial solvents and cleaning agents, and their widespread use and improper disposal practices have led to them becoming common groundwater contaminants throughout the USA and the world [25, 33]. Because of the widespread occurrence of chlorinated solvent contamination, a number of treatment technologies have emerged and evolved. Currently, the most common treatment alternative involves biological degradation of the solvents.
The predominant biodegradation pathway for chlorinated ethenes under anaerobic conditions is reductive dechlorination. During reductive dechlorination, chlorinated ethenes are used as electron acceptors by specialized microorganisms, and during the process a chlorine atom is removed and replaced with a hydrogen atom [12, 13, 16, 30]. Sequential dechlorination of perchloroethylene (PCE) most commonly proceeds to trichloroethene (TCE), cis-1,2-dichloroethene (cDCE), vinyl chloride (VC), and finally the desired end product, ethene. In some cultures trans-1,2-DCE and 1,1-DCE also can be produced through the reductive dechlorination of TCE [6, 35]. In situ biodegradation of chlorinated ethenes can be performed by indigenous microorganisms at contaminated sites that use endogenous resources to support contaminant degradation (i.e., intrinsic bioremediation), or nutrients that are purposefully added to support their activity (i.e., biostimulation). The lack of an adequate microbial population capable of completely dechlorinating PCE and TCE to ethene at some sites, however, may lead to the accumulation of cis-DCE and VC [11]. Consequently, the addition of exogenous organisms (i.e., bioaugmentation) is sometimes used to supplement the indigenous microbial population [5, 15, 21].
While many dechlorinating microorganisms have been identified [30], bacteria of only one microbial genus, Dehalococcoides (DHC), have been shown to completely reduce cDCE and VC to ethene [7, 8, 22, 23, 26, 31]. These organisms use molecular hydrogen as an obligate electron donor and halogenated compounds as obligate respiratory electron acceptors. Acetate (e.g., from lactate fermentation) is used as a carbon source. Studies of field sites have strongly correlated the presence of DHC strains with complete dehalogenation of chlorinated ethenes in situ [11]. Therefore, microbial cultures used to remediate chlorinated solvent-contaminated groundwaters contain at least one strain of Dehalococcoides sp. Because of the difficulty of growing DHC-type organisms in pure culture [7, 8, 10, 23], however, cultures used for bioaugmentation applications are consortia that contain DHC as well as fermentative and other microbes that support the growth and activity of the DHC strains [4, 5, 15, 21]. The consortia, and the DHC therein, can be grown on a wide range of carbon sources provided the substrate is fermented to hydrogen.
One of the significant challenges of performing bioaugmentation at a commercial scale is the large size of contaminant plumes and the large amount of culture needed to facilitate timely and successful remediation. Contaminant plumes can range from less than an acre (0.4 ha) in size to several kilometers long and hundreds of meters wide. Recent studies of in situ chlorinated ethene degradation have suggested that DHC concentrations in the range of 107 DHC/L of groundwater are needed to support acceptable degradation rates [19, 28]. To illustrate the challenge of applying bioaugmentation in the field, a 0.4-ha (one-acre) aquifer with a saturated zone 3 m (10 ft) thick and porosity of 25% would contain ~3 × 106 L of groundwater and require 3 × 1013 DHC based on the findings of Lu et al. At the reported DHC concentrations of early bioaugmentation cultures (109 DHC/L; [21]), as much as 104 L of culture could be required to treat a one-acre site. Of course other factors affect the amount of culture applied at a site [14, 28], but it is clear that large-scale production of high-density cultures is necessary to apply bioaugmentation economically, especially at large sites.
The objective of this study is to evaluated large-scale production of a DHC-containing consortium, SDC-9™, for full-scale remedial applications. The culture was grown in small (3-L) to large (4,000-L) fermentors by using sodium lactate as a carbon and electron donor source and PCE as an electron acceptor. DHC concentrations of >1011/L could be achieved, and the culture could be concentrated and stored prior to field application. The fermentation procedure produced similar results with two other DHC cultures enriched from different sites.
Materials and methods
Chemicals
Sodium-(l)-lactate (60% solution) was purchased from Purac America (Lincolnshire, IL), yeast extract (bacteriological grade) was purchased from Marcor Development Corp. (Carlstadt, NJ), and PCE (99.9%) was from Sigma/Aldrich (Milwaukee, WI). Unless otherwise stated, all other chemicals were of the highest purity available and purchased from either Aldrich Chemical Co. (Milwaukee, WI), Mallinckrodt Specialty Chemical Co. (Paris, KY), J.T. Baker Inc. (Phillipsburg, NJ.), Spectrum Chemical Manufacturing Corp. (Garden, CA) or Sigma Chemical Co. (St. Louis, MO).
Bacterial cultures
An anaerobic dechlorinating consortium designated SDC-9™ was isolated by enrichment culturing of samples from a chlorinated solvent-contaminated aquifer in southern California with lactate as an electron donor and PCE as an electron acceptor. The culture has been maintained on sodium lactate and PCE in reduced anaerobic mineral medium (RAMM) [29], but without sodium sulfide and rezasurin, for more than 4 years. Hawaii-05™ was enriched in 2005 by enrichment culturing of aquifer samples from Hickam Air Force Base, Hawaii on sodium lactate and TCE, and PJKS™ was enriched in 2005 from aquifer samples from Air Force Plant PJKS in Colorado on sodium lactate and TCE. The latter cultures are maintained as described for SDC-9™. All three cultures are marketed commercially by Shaw Environmental, Inc. (Lawrenceville, NJ).
Fermentation equipment
Bench-scale fermentation experiments and seed culture production were performed in a 3-L or 7-L Applicon fermentor (Cole Palmer, Vernon Hills, IL.) equipped with pH and mixer controls. Substrate and NaOH feeds were controlled by using syringe pumps (Harvard Apparatus, Holliston, MA). Larger seed cultures were produced in a similarly equipped 20-L Biolafitte fermentor (Pierre Guerin, Inc., Spring Lake Park, MN). Larger cultures were produced in a 750-L ABEC fermentor (Bethlehem, PA) or a custom-built 4,000-L stainless-steel fermentor. In each case anaerobic conditions were maintained by pressurizing the vessels with nitrogen. Cells in the fermentation broth were concentrated by passing the broth over a custom-built concentrator constructed with six Kerasep™ tubular ceramic membranes (Novasep, Inc., Boothwyn, PA). Concentrated cells were stored at 4°C in 18.5-L stainless-steel soda kegs that were pressurized with nitrogen.
Fermentation protocol
For seed culture production RAMM medium [29] without NaHCO3 and Na2S was added to the 20-L fermentor and steam sterilized at 121°C and 15 psi pressure for 45 min. After sterilization the fermentor was connected to a nitrogen tank to maintain a positive pressure of nitrogen in the fermentor during cooling to 30°C. After the temperature in the fermentor reached the set-point temperature of fermentation (28–30°C) and anaerobic condition were achieved [measured dissolved oxygen (DO) = 0 mg/L], nitrogen flow was stopped and NaHCO3 solution was added aseptically to the medium. The fermentor was then inoculated with 2 L of SDC-9™, PJKS™ or Hawaii-05™. The final volume of medium in the fermentor was 16–18 L.
After inoculation of the fermentor, sterile 10% yeast extract (YE) solution was added to a final concentration of 0.1% YE (w/v) and PCE or TCE was added to a final concentration of 10 mg/L. SDC-9™ was grown on PCE, but PJKS™ and Hawaii-05™ were grown on either PCE or TCE. The fermentor was operated at 28–30°C with an agitator speed of 100 rpm. pH was maintained at 6.4–7.2 by addition NaOH (2 N). Alternatively, to increase pH during fermentation, the fermentor was sparged with nitrogen to remove dissolved CO2. To control foam in the fermentor Antifoam 289 or 204 (Sigma) was applied automatically. After 1 day of fermentation, sodium lactate (60% solution) was added continuously to the fermentor at flow rate of 0.02–0.04 mL/h × liter of media. Subsequent additions of PCE or TCE (10 mg/L) were made to the fermentor only after complete dechlorination of PCE/TCE but before complete dechlorination of cDCE. Typically, PCE/TCE was added to the medium when the concentration of cDCE in the medium was reduced to 1–3 mg/L. When the culture reached an optical density (OD) at 550 nm (OD550) of approximately 1.0 it was transferred anaerobically to the 750-L fermentor.
The 750-L fermentor was prepared with 550 L RAMM medium and sampled and monitored essentially as described above. The fermentor was connected to a nitrogen tank to maintain anoxic conditions, and it was operated under the same conditions as described for the 20-L fermentor except the agitator speed was set at 60 rpm. The automatic pH control system on the fermentor was inactivated to avoid addition of excess sodium. After 1 day of fermentation a continuous feed of sodium lactate (60% solution) was initiated with flow rate of 0.02–0.04 mL/h × L. When the specific PCE and cDCE dechlorination activity reached 1.3–1.7 mg/h × gram of dry weight, a continuous feed of neat PCE/TCE was initiated at rate of 0.18–0.25 μL/h × L. This rate was increased to 0.9–1.2 μL/h × L as the culture cell density and dechlorination activity increased. The culture was grown for 13–15 days until an OD550 ≈ 0.7–1.1 or 1010-1011 DHC/L was achieved. Higher DHC concentrations could be obtained by extending the fermentation for up to 35 days.
Growth of the cultures in the 4,000-L fermentor (working volume 3,200 L) was performed essentially as described for the 750-L fermentor, but because the 4,000-L fermentor did not have an impeller, cells were continuously suspended by using a centrifugal pump that circulated the culture medium. The 4,000-L fermentor was chemically sterilized by using NaOH and a clean-in-place system. The culture medium in the 4,000-L fermentor was not sterilized. Substrate feeding and other parameters were as described for the 750-L fermentor. The fermentor was inoculated with either culture from the 750-L fermentor or refrigerated concentrated cell stocks.
Degradation assays and analytical procedures
Whenever possible, analytical methods performed during this project followed US Environmental Protection Agency (USEPA) SW-846 methods [32] that are available online at http://www.epa.gov/epawaste/hazard/testmethods/sw846/index.htm. Biodegradation assays were incubated at 28 ± 1°C in the dark in serum vials essentially as described by Schaefer et al. [28]. Chlorinated ethene analyses were performed by gas chromatography using USEPA method 8260 [gas chromatography/mass spectrometry (GC/MS) with purge and trap injection]. Methane and ethene were monitored by GC/flame ionization detection (FID) according to USEPA SW846 method 8015b. Lactate and volatile fatty acids (VFAs) were measured by ion chromatography using USEPA method 300.0-modified on a Dionex DX600 ion chromatograph (Dionex Corp., Bannockburn, IL). Hydrogen concentration in the fermentors was measured by analyzing the headspace of 100-mL samples in 120-mL vials on a Varian 3800 gas chromatograph (Varian, Inc., Walnut Creek, CA) equipped with a Valco pulsed discharge helium ionization detector (PDHID), a helium gas purifier to achieve helium carrier and makeup gas of 99.999% purity, and Varian Pora Bond Q (10 m, 0.32 inner diameter, 5 uM df) and Varian Molsieve 5A (10 m, 0.32 inner diameter, 5 µM df) columns operated in series. Concentration of hydrogen was determined by comparison to a standard curve. Dry weight (Dwt) was determined by concentrating 15–30 mL culture in a RC5C centrifuge (10,000×g; Sorval Instruments, Newtown, CT), removing the supernatant, suspending the pellet in deionized (DI) water, and repeating the procedure twice. The washed cell pellet was suspended in DI water, transferred to an aluminum weighing dish, and dried at 105°C.
DHC quantification
DHC-like organisms were quantified by using real-time quantitative polymerase chain reaction (qPCR). Following collection of fermentor samples, the OD550 of the sample was measured and the cells were either concentrated by centrifugation or diluted with water to an OD550 of approximately 0.5. OD was then remeasured for verification. One milliliter of the OD550 = 0.5 cells were then concentrated by centrifugation (16,000×g for 2 min) and resuspended in 100 μL distilled water. The cells were then processed using an Idaho Technologies 1-2-3 RAPID DNA purification kit (Idaho Technology Inc. Salt Lake City, UT) as per manufacturer instructions and using a Bead Beater (BioSpec Products Inc., Tulsa, OK). DNA was eluted from columns in a final volume of 100 μL buffer rather than the prescribed 400 μL.
Quantitative real-time PCR was performed with a RAPID PCR machine (Idaho Technologies Inc.) and a Lightcycler FastStart DNA Master Hybprobe probe kit (Roche Diagnostics GmbH, Manheim, Germany) and primers developed by us with the assistance of Idaho Technologies, Inc. to amplify and quantify 16 s ribosomal RNA (rRNA) gene DNA. DNA amplification used a forward primer (5′-GAAGTAGTGAACCGAAAGG-3′) and a reverse primer (5′-TCTGTCCATTGTAGCGTG-3′), and the amplified DNA was quantified using a fluorescence resonance energy transfer (FRET) probe system that employed a Light Cycler Red 640 fluorophore (5′-AGCGAGACTGCCCC-3′) and an fluorescein isothiocyanate (FITC)-labeled probe (5′-CCCACCTTCCTCCCCGTTTC-3′). The amplification conditions were as follows: denaturation at 95°C for 10 min, followed by 40 cycles of melting at 94°C for 20 s, annealing at 53°C for 10 s, and extension at 72°C for 20 s. Dehalococcoides sp. chromosomal DNA was quantified by comparison to a standard curve generated by amplifying serial dilutions of a known concentration of plasmid (pSC-A vector; Stratagene Inc. La Jolla, CA) containing a cloned 16S rRNA gene from the SDC-9™ culture.
Results and discussion
Culture growth
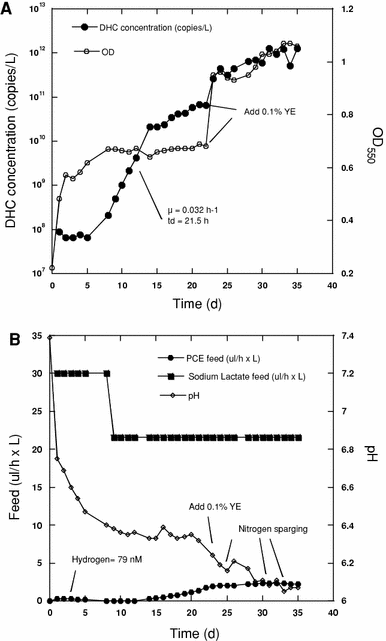
Growth of SDC-9™ in a 4,000-L fermentor. a Concentration of DHC as measured by qPCR (filled circle) and total cell concentration as estimated by OD at 550 nm (open circle). DHC first-order growth rate (μ) and doubling time (td) are indicated on the graph. b Feed rate of neat PCE (filled circle) and 60% sodium lactate (filled square), and the pH of the culture medium (open diamond) are indicated. Yeast extract (YE) solution was added at the beginning of the fermentation and as indicated. The fermentor was sparged with N2 as indicated to control pH
Although the OD of the culture stabilized after approximately 10 days, exponential growth of DHC continued until approximately day 24. These results suggest that non-DHC microorganisms in the consortium initially grew much faster than DHC. During this early fermentation period, DHC represented a relatively low proportion of the total bacterial population of the culture, but during extended growth the relative abundance of DHC in the culture increased. The results also demonstrate that, at least during the early stages of fermentation, OD measurements are not a good indicator of DHC concentration in the culture, and more advanced measurements such as qPCR are needed to estimate DHC numbers in the culture effectively [17, 27].
During the initial stages of 3,200-L fermentation (to day 25) a maximum DHC concentration of ~1011 DHC/L was achieved in the fermentor, even though growth substrates were still present in the culture broth (Fig. 1a). DHC concentrations in the fermentor, however, could be increased approximately tenfold by the addition of YE as a nutrient source. The exact role of the YE is not known, but its addition also revived the growth of non-DHC organisms in the consortium (Fig. 1a). Because the RAMM medium used in this study did not contain sodium sulfide or other sulfur-containing salts, it is possible that the yeast extract provided a needed source of sulfur for the cultures. Based on our analysis (data not shown) 1 g/L YE was estimated to provide 5 mg/L sulfur and 0.48 mg/L iron. YE also could provide a needed source of amino acids and/or precursors for the production of corrinoid cofactors that are necessary for dehalogenation by DHC strains [23]. During this extended growth of the culture there was a correlation between culture OD550 and DHC concentrations, suggesting that during this period of the fermentation process measurements of OD may be useful for estimating DHC levels in the fermentor and to automate the control of the fermentation process.
Similar fermentation results were obtained with two other chloroethene dechlorinating bacterial consortia, PJKS™ and Hawaii-05™, at both the 550-L and 3,200-L scale (Table 1), by using the described procedures. Both cultures could be grown to high DHC concentration (>1011 cells/L), and both the final OD550 and total cell mass obtained were similar to the results obtained with SDC-9™.
Results of multiple fermentation runs with the tested chlorinated solvent-dechlorinating consortia
| Culture . | Date (M/Y) . | Volume (L) . | Final OD550 . | Final DHC (cells/L)a . | Dwt (mg/L) . | PCE activity (mg/h/g Dwt) . | cDCE activity (mg/h/g Dwt) . |
|---|---|---|---|---|---|---|---|
| SDC-9 | 01/2006 | 550 | 1.3 | 1.4 E11 | 0.51 | 16 | 13 |
| SDC-9 | 02/2008 | 550 | 1.7 | 2.8 E11 | 0.66 | 22 | 14 |
| SDC-9 | 03/2008 | 3,200 | 1.6 | 1.4 E11 | 0.65 | 41 | 37 |
| SDC-9 | 05/2008 | 2,500 | 1.6 | 2.4 E12 | 0.59 | 42 | 39 |
| SDC-9 | 08/2008 | 2,000 | 1.4 | 1.0 E12 | 0.51 | 80 | 69 |
| PJKS | 01/2008 | 2,500 | 1.1 | 9.4 E11 | 0.41 | 32 | 14 |
| PJKS | 02/2008 | 1,700 | 1.3 | 1.0 E11 | 0.50 | 64 | 45 |
| Hawaii-05 | 11/2007 | 550 | 1.2 | 1.5 E11 | 0.50 | 23 | 16 |
| Culture . | Date (M/Y) . | Volume (L) . | Final OD550 . | Final DHC (cells/L)a . | Dwt (mg/L) . | PCE activity (mg/h/g Dwt) . | cDCE activity (mg/h/g Dwt) . |
|---|---|---|---|---|---|---|---|
| SDC-9 | 01/2006 | 550 | 1.3 | 1.4 E11 | 0.51 | 16 | 13 |
| SDC-9 | 02/2008 | 550 | 1.7 | 2.8 E11 | 0.66 | 22 | 14 |
| SDC-9 | 03/2008 | 3,200 | 1.6 | 1.4 E11 | 0.65 | 41 | 37 |
| SDC-9 | 05/2008 | 2,500 | 1.6 | 2.4 E12 | 0.59 | 42 | 39 |
| SDC-9 | 08/2008 | 2,000 | 1.4 | 1.0 E12 | 0.51 | 80 | 69 |
| PJKS | 01/2008 | 2,500 | 1.1 | 9.4 E11 | 0.41 | 32 | 14 |
| PJKS | 02/2008 | 1,700 | 1.3 | 1.0 E11 | 0.50 | 64 | 45 |
| Hawaii-05 | 11/2007 | 550 | 1.2 | 1.5 E11 | 0.50 | 23 | 16 |
aBased on qPCR assuming 1 16S rRNA gene copy/cell
Results of multiple fermentation runs with the tested chlorinated solvent-dechlorinating consortia
| Culture . | Date (M/Y) . | Volume (L) . | Final OD550 . | Final DHC (cells/L)a . | Dwt (mg/L) . | PCE activity (mg/h/g Dwt) . | cDCE activity (mg/h/g Dwt) . |
|---|---|---|---|---|---|---|---|
| SDC-9 | 01/2006 | 550 | 1.3 | 1.4 E11 | 0.51 | 16 | 13 |
| SDC-9 | 02/2008 | 550 | 1.7 | 2.8 E11 | 0.66 | 22 | 14 |
| SDC-9 | 03/2008 | 3,200 | 1.6 | 1.4 E11 | 0.65 | 41 | 37 |
| SDC-9 | 05/2008 | 2,500 | 1.6 | 2.4 E12 | 0.59 | 42 | 39 |
| SDC-9 | 08/2008 | 2,000 | 1.4 | 1.0 E12 | 0.51 | 80 | 69 |
| PJKS | 01/2008 | 2,500 | 1.1 | 9.4 E11 | 0.41 | 32 | 14 |
| PJKS | 02/2008 | 1,700 | 1.3 | 1.0 E11 | 0.50 | 64 | 45 |
| Hawaii-05 | 11/2007 | 550 | 1.2 | 1.5 E11 | 0.50 | 23 | 16 |
| Culture . | Date (M/Y) . | Volume (L) . | Final OD550 . | Final DHC (cells/L)a . | Dwt (mg/L) . | PCE activity (mg/h/g Dwt) . | cDCE activity (mg/h/g Dwt) . |
|---|---|---|---|---|---|---|---|
| SDC-9 | 01/2006 | 550 | 1.3 | 1.4 E11 | 0.51 | 16 | 13 |
| SDC-9 | 02/2008 | 550 | 1.7 | 2.8 E11 | 0.66 | 22 | 14 |
| SDC-9 | 03/2008 | 3,200 | 1.6 | 1.4 E11 | 0.65 | 41 | 37 |
| SDC-9 | 05/2008 | 2,500 | 1.6 | 2.4 E12 | 0.59 | 42 | 39 |
| SDC-9 | 08/2008 | 2,000 | 1.4 | 1.0 E12 | 0.51 | 80 | 69 |
| PJKS | 01/2008 | 2,500 | 1.1 | 9.4 E11 | 0.41 | 32 | 14 |
| PJKS | 02/2008 | 1,700 | 1.3 | 1.0 E11 | 0.50 | 64 | 45 |
| Hawaii-05 | 11/2007 | 550 | 1.2 | 1.5 E11 | 0.50 | 23 | 16 |
aBased on qPCR assuming 1 16S rRNA gene copy/cell
No other studies have evaluated or reported large-scale production of DHC-containing consortia, but the DHC cell concentration achieved in our studies were similar to those obtained by others in small-scale laboratory tests. For example, Couples et al. [1] calculated final DHC concentrations of up to 4 × 1011/L during growth of the VS culture in TCE-fed 60-mL batch cultures, and He et al. [9], achieved up to 1.8 × 1011 copies/L of the tceA gene in 100-mL batch cultures of D. ethenogenes strain 195 containing a coculture of a sulfate-reducing bacterium. Similarly, whereas we observed DHC doubling times of 19.3 h during large-scale fermentation, DHC doubling times from small laboratory studies of 19.5 h to 2 days have been reported [2, 9, 10, 22].
The results of this study demonstrate that culture volumes and DHC cell densities sufficient to treat even relatively large contaminated aquifers can be obtained. Assuming that 107 DHC/L of contaminated groundwater are needed to obtain effective and timely remediation [19], 3,200 L of culture with 1011 DHC/L could potentially support remediation of 3.2 × 107 L of groundwater, even without further in situ growth of the organisms.
Factors affecting fermentation
Several factors could affect the results obtained during growth of the test cultures, including substrate type and feed rates, pH, and VFA accumulation. Growth of DHC requires the presence of a chlorinated substrate as an electron acceptor, H2 as an electron donor, and a carbon growth source such as acetate [8, 16, 23]. In consortia such as those used in this study, the primary growth substrate (i.e., lactate) is fermented by non-DHC members of the consortia to H2 and acetate that can be utilized by DHC. The presence of excess H2, however, can lead to substrate competition with methanogenic bacteria in the consortia that also can use H2, albeit at a higher substrate threshold than DHC [18, 20, 34]. Therefore, in developing a fermentation protocol for the described cultures, attempts were made to maintain consistent low H2 concentrations within the reactor. The sodium lactate feed rate used during the fermentation process resulted in sustained dissolved hydrogen concentration in the reactor of <20 nM. During utilization of the initial batch feeding of lactate and YE added prior to inoculation, H2 concentrations sometimes exceeded 100 nM, but during the extended fermentation process H2 concentrations were typically 3–5 nM, which was similar to the half-velocity coefficient for hydrogen calculated for the VS culture (7 ± 2 nM; [3]).
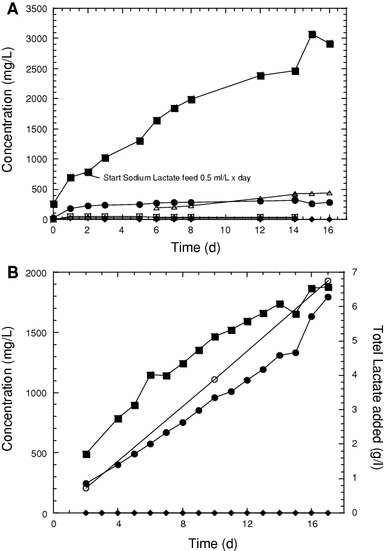
Accumulation of VFAs during growth of SDC-9™ (a) or PJKS™ (b) in a 750-L fermentor. Symbols indicate lactic acid (filled diamond), propionic acid (filled circle), formic acid (open diamond), pyruvic acid (open square), butyric acid (open triangle), and acetic acid (filled square), or the total amount of sodium lactate added to the fermentor (open circle; b)
To optimize the growth of the SDC-9™ consortium it was necessary to determine a relationship between PCE feed rate and DHC cell concentration. We were most concerned about maintaining the VC-reducing population(s) in the consortia because VC reduction is less energetically favorable than the other dehalogenating reactions, so it was possible that PCE and TCE dehalogenating populations could outcompete the VC reducers if the higher chlorinated substrates were maintained in excess. Furthermore, Cupples et al. [3] observed that net decay in dechlorinating microorganisms could occur in the VS culture if DCE plus VC concentrations were below 0.7 μM. In addition, with SDC-9™, based on many biodegradation assays, the VC dechlorination rate is 28–35% of the PCE dechlorination rate. Therefore, there was a tendency for VC to accumulate in the fermentor during high-rate PCE feeding. Consequently, PCE feed rates were adjusted to prevent accumulation of PCE, TCE or cis-DCE while maintaining a residual VC concentration in the medium of ~1 mg/L (16 μM). Evaluating the PCE feed rates during multiple fermentation runs, the results of the biodegradation assays, and the analyses of PCE, TCE cDCE, and VC concentrations during fermentation allowed us to optimize PCE feed rates for the growth of SDC-9™ consortium. The relationship between DHC yield and PCE feed rate could be described by the following equation: DHC concentration (cells/L) = −6.77 × 1011 + [8.40 × 1011 × PCE feed rate (mg/h × L)] (R = 0.999).
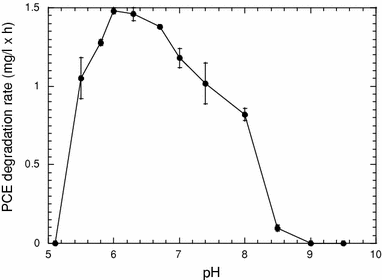
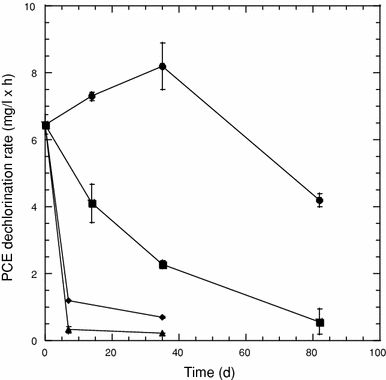
Effect of culture pH on PCE dehalogenation by SDC-9™. Values represent the mean of triplicate samples, and error bars represent one standard error of the mean
Culture activity
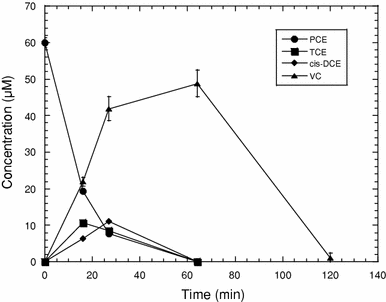
Results of a PCE degradation assay with samples from a 550-L fermentation batch of the SDC-9™ culture. The assay was performed in 60-mL serum vials containing 60 mL SDC-9™ culture (0.52 g/L Dwt), 6 mM sodium lactate, and 10 mg/L PCE. Values represent means of triplicate samples and error bars represent one standard error of the mean
Related issues
The use of bioaugmentation to remediate chlorinated solvent-contaminated sites requires the shipment of cultures throughout the USA and elsewhere. Shipping a large volume of culture is costly, and ground transportation can require that the culture spend several days in shipping, which could affect culture activity. An alternate approach is to concentrate the culture to allow overnight shipping of a reduced culture volume. We used a tubular ceramic membrane system to concentrate consortia. The cell culture was chilled during concentration to ensure maintenance of cell viability. Analysis of the specific activity of the cells before and after concentration demonstrated only slight changes in activity during concentration. For example, specific activity of two cultures tested were 24.5 and 16.5 mg PCE/h × g Dwt before concentration and 22.6 and 15.1 mg PCE/h × g Dwt after concentration, respectively. Concentration resulted in approximately 90% reduction in culture volume, and it also removed ~90% of any fermentation byproducts remaining in the culture broth. It also allowed us to standardize the DHC concentration and activity of culture batches, thereby allowing users to more accurately estimate the volume of culture needed for field applications.
Effect of storage conditions on the activity of concentrated SDC-9™ culture. Tenfold-concentrated SDC-9™ culture was stored anaerobically and without substrate at either 4°C (filled circle), 13°C (filled square), 22°C (filled diamond) or 28°C (filled triangle). Values represent means of triplicate samples, and error bars are one standard error of the mean
Conclusions
A fermentation protocol was developed for large-scale production of DHC-containing cultures for in situ bioaugmentation of chlorinated ethene-contaminated aquifers. The performance of the SDC-9™ culture in contaminated aquifer material is described elsewhere [28]. Success of the fermentation process was dependant on electron donor (i.e., lactate) and acceptor (PCE) feed rate, and the addition of YE greatly improved cell yield. The initial stages of fermentation were characterized by a rapid growth of non-DHC organisms in the culture, while the growth rate of DHC within the consortia tested exhibited a short lag and then was relatively constant to final DHC concentrations of >1011/L. The fermentation protocol was scalable to 550 L and 3,200 L and produced comparable results for consortia enriched from three different sites.
Based on 16S RNA gene sequencing the SDC-9™ culture contains multiple DHC strains (data not shown), and it is possible that growth of the individual dehalogenating strains within the culture might be different during the fermentation process. Although this could not be monitored during this study, our results demonstrated that both PCE and cDCE dehalogenation activities were high in the final cultures, and the culture degraded VC well, albeit at a lower rate than PCE and cDCE dehalogenation. This suggests that the described procedure supports the growth of DHC that are able to completely dehalogenate chlorinated ethenes, including vinyl chloride. Our results also demonstrate that DHC-containing cultures designed for bioaugmentation can be concentrated by cross-flow filtration to reduce shipping volumes, and that the concentrated cultures can be stored under refrigeration for >40 days to allow for injection schedule flexibility.
With the increased use of bioaugmentation to treat challenging chlorinated ethene-contaminated sites, the ability to produce large volumes of high-density cultures is becoming increasingly important. This study provides useful information to aid in the production of cultures for bioaugmentation, even at scales suitable for treating large contaminant plumes.
Acknowledgments
The authors thank Randi Rothmel, Antonio Soto, Kevin McClay, and Paul Hedman for excellent analytical support. This project was supported by the Environmental Security Technology Certification Program (ESTCP) project number CU-0515. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect those of the US Army Corp. of Engineers, Humphreys Engineer Center Support Activity.
References
Löffler FE, Cole JR, Ritalahti KM, Tiedje JM (2003) Diversity of dechlorinating bacteria. In: Häggblom MM, Bossert ID (eds) Dehalogenation: microbial processes and environmental applications. Kluwer Academic Press, New York, pp 53–87. doi: 10.1007/0-306-48011-5_3
U.S. EPA (1998) U.S. EPA test methods for evaluating solid waste, physical/chemical methods SW846, 3rd edn. Revision 5, 1998