Abstract
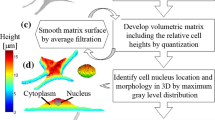
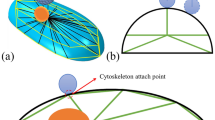
The goal of this study is to construct a representative 3D finite element model (FEM) of individual cells based on their sub-cellular structures that predicts cell mechanical behavior. The FEM simulations replicate atomic force microscopy (AFM) nanoindentation experiments on live vascular smooth muscle cells. Individual cells are characterized mechanically with AFM and then imaged in 3D using a spinning disc confocal microscope. Using these images, geometries for the FEM are automatically generated via image segmentation and linear programming algorithms. The geometries consist of independent structures representing the nucleus, actin stress fiber network, and cytoplasm. These are imported into commercial software for mesh refinement and analysis. The FEM presented here is capable of predicting AFM results well for 500 nm indentations. The FEM results are relatively insensitive to both the exact number and diameter of fibers used. Despite the localized nature of AFM nanoindentation, the model predicts that stresses are distributed in an anisotropic manner throughout the cell body via the actin stress fibers. This pattern of stress distribution is likely a result of the geometric arrangement of the actin network.










Similar content being viewed by others
Abbreviations
- AFM:
-
Atomic force microscope
- CytoD:
-
Cytochalasin D
- FEM:
-
Finite element model
- ST:
-
Simulation type
- VSMC:
-
Vascular smooth muscle cell
References
Athanasiou, K. A., B. S. Thoma, D. R. Lanctot, D. Shin, C. M. Agrawal, and R. G. LeBaron. Development of the cytodetachment technique to quantify mechanical adhesiveness of the single cell. Biomaterials 20(23–24):2405–2415, 1999.
Balogh, J., M. Merisckay, Z. Li, D. Paulin, and A. Arner. Hearts from Mice Lacking Desmin have a myopathy with impaired active force generation and unaltered wall compliance. Cardiovasc. Res. 53(2):439–450, 2002.
Buerke, M., M. Guckenbiehl, H. Schwertz, U. Buerke, M. Hilker, H. Platsch, J. Richert, S. Bomm, G. A. Zimmerman, S. Lindemann, U. Mueller-Werdan, K. Werdan, H. Darius, and A. S. Weyrich. Intramural delivery of sirolimus prevents vascular remodeling following balloon injury. Biochim. Biophys. Acta 1774(1):5–15, 2007.
Casscells, W. Migration of smooth muscle and endothelial cells—critical events in restenosis. Circulation 86(3):723–729, 1992.
Chicurel, M. E., C. S. Chen, and D. E. Ingber. Cellular control lies in the balance of forces. Curr. Opin. Cell Biol. 10(2):232–239, 1998.
Costa, K. D., W. J. Hucker, and F. C. P. Yin. Buckling of Actin stress fibers: a new wrinkle in the cytoskeletal tapestry. Cell Motil. Cytoskeleton. 52(4):266–274, 2002.
Dailey, H. L., L. M. Ricles, H. C. Yalcin, and S. N. Ghadiali. Image-based finite element modeling of alveolar epithelial cell injury during airway reopening. J. Appl. Physiol. 106(1):221–232, 2009.
Gittes, F., B. Mickey, J. Nettleton, and J. Howard. Flexural rigidity of microtubules and actin filaments measured from thermal fluctuations in shape. J. Cell Biol. 120(4):923–934, 1993.
Gustafsson, M. G. L. Extended resolution fluorescence microscopy. Curr. Opin. Struct. Biol. 9(5):627–628, 1999.
Hemmer, J. D., D. Dean, A. Vertegel, E. Langan, III, and M. LaBerge. Effects of serum deprivation on the mechanical properties of adherent vascular smooth muscle cells. Proc. Inst. Mech. Eng. H 222(5):761–772, 2008.
Hemmer, J. D., J. Nagatomi, S. T. Wood, A. A. Vertegel, D. Dean, and M. LaBerge. Role of cytoskeletal components in stress-relaxation behavior of adherent vascular smooth muscle cells. J. Biomech. Eng. 131(4):9, 2009.
Ingber, D. Integrins as mechanochemical transducers. Curr. Opin. Cell Biol. 3(5):841–848, 1991.
Ingber, D. E. Mechanobiology and diseases of mechanotransduction. Ann. Med. 35(8):564–577, 2003.
Keller, R. S., S. Y. Shai, C. J. Babbitt, C. G. Pham, R. J. Solaro, M. L. Valencik, J. C. Loftus, and R. S. Ross. Disruption of integrin function in the murine myocardium leads to perinatal lethality, fibrosis, and abnormal cardiac performance. Am. J. Pathol. 158(3):1079–1090, 2001.
Kojima, H., A. Ishijima, and T. Yanagida. Direct measurement of stiffness of single actin filaments with and without tropomyosin by in vitro nanomanipulation. Proc. Natl. Acad. Sci. USA 91(26):12962–12966, 1994.
Kozubek, M., S. Kozubek, E. Lukasova, A. Mareckova, E. Bartova, M. Skalnikova, and A. Jergova. High-resolution cytometry of fish dots in interphase cell nuclei. Cytometry 36(4):279–293, 1999.
Kumar, S., I. Z. Maxwell, A. Heisterkamp, T. R. Polte, T. P. Lele, M. Salanga, E. Mazur, and D. E. Ingber. Viscoelastic retraction of single living stress fibers and its impact on cell shape, cytoskeletal organization, and extracellular matrix mechanics. Biophys. J. 90(10):3762–3773, 2006.
Li, C., and Q. Xu. Mechanical stress-initiated signal transduction in vascular smooth muscle cells in vitro and in vivo. Cell. Signal. 19(5):881–891, 2007.
Li, G., T. M. Liu, A. Tarokh, J. X. Nie, L. Guo, A. Mara, S. Holley, and S. T. C. Wong. 3D cell nuclei segmentation based on gradient flow tracking. BMC Cell Biol. 8:40, 2007.
Li, T. A mechanics model of microtubule buckling in living cells. J. Biomech. 41(8):1722–1729, 2008.
Lin, G., U. Adiga, K. Olson, J. F. Guzowski, C. A. Barnes, and B. Roysam. A hybrid 3D watershed algorithm incorporating gradient cues and object models for automatic segmentation of nuclei in confocal image stacks. Cytometry A. 56(1):23–36, 2003.
Lin, G., M. K. Chawla, K. Olson, J. F. Guzowski, C. A. Barnes, and B. Roysam. Hierarchical, model-based merging of multiple fragments for improved three-dimensional segmentation of nuclei. Cytometry A J. Int. Soc. Analyt. Cytol. 63A(1):20–33, 2005.
Loufrani, L., K. Matrougui, Z. L. Li, B. I. Levy, P. Lacolley, D. Paulin, and D. Henrion. Selective microvascular dysfunction in mice lacking the gene encoding for desmin. FASEB J. 15(13):117–119, 2001.
Luo, Y., X. Xu, T. Lele, S. Kumar, and D. E. Ingber. A multi-modular tensegrity model of an actin stress fiber. J. Biomech. 41(7):2379–2387, 2008.
McManus, K. J., D. A. Stephens, N. M. Adams, S. A. Islam, P. S. Freemont, and M. J. Hendzel. The transcriptional regulator CBP has defined spatial associations within interphase nuclei. PLoS Comput. Biol. 2(10):1271–1283, 2006.
Melder, R. J., C. A. Kristensen, L. L. Munn, and R. K. Jain. Modulation of A-NK cell rigidity: in vitro characterization and in vivo implications for cell delivery. Biorheology. 38(2–3):151–159, 2001.
Mofrad, M. R. K. and R. D. Kamm (eds.). Cytoskeletal Mechanics. New York, NY: Cambridge University Press, p. 244, 2006.
MSC, S.C. When F =/= Ku, pp. 2–11, 2009.
Nagayama, K., Y. Yahiro, and T. Matsumoto. Stress fibers stabilize the position of intranuclear DNA through mechanical connection with the nucleus in vascular smooth muscle cells. FEBS Lett. 585(24):3992–3997, 2011.
Park, C. Y., D. Tambe, A. M. Alencar, X. Trepat, E. H. Zhou, E. Millet, J. P. Butler, and J. J. Fredberg. Mapping the cytoskeletal prestress. Am. J. Physiol. Cell Physiol. 298(5):C1245–C1252, 2010.
Peeters, E. A. G., C. W. J. Oomens, C. V. C. Bouten, D. L. Bader, and F. P. T. Baaijens. Mechanical and failure properties of single attached cells under compression. J. Biomech. 38(8):1685–1693, 2005.
Pullarkat, P. A., P. A. Fernández, and A. Ott. Rheological properties of the eukaryotic cell cytoskeleton. Phys. Rep. 449(1–3):29–53, 2007.
Russell, R. A., N. M. Adams, D. A. Stephens, E. Batty, K. Jensen, and P. S. Freemont. Segmentation of fluorescence microscopy images for quantitative analysis of cell nuclear architecture. Biophys. J. 96(8):3379–3389, 2009.
Shiels, C., S. A. Islam, R. Vatcheva, P. Sasieni, M. J. E. Sternberg, P. S. Freemont, and D. Sheer. PML bodies associate specifically with the MHC gene cluster in interphase nuclei. J. Cell Sci. 114(20):3705–3716, 2001.
Slomka, N., and A. Gefen. Confocal microscopy-based three-dimensional cell-specific modeling for large deformation analyses in cellular mechanics. J. Biomech. 43(9):1806–1816, 2010.
Unnikrishnan, G. U., V. U. Unnikirishnan, and J. N. Reddy. Constitutive material modeling of cell: a micromechanics approach. J. Biomech. Eng. 129(3):315–323, 2007.
Wang, J., C. Shiels, P. Sasieni, P. J. Wu, S. A. Islam, P. S. Freemont, and D. Sheer. Promyelocytic leukemia nuclear bodies associate with transcriptionally active genomic regions. J. Cell Biol. 164(4):515–526, 2004.
Waters, C. M., P. H. S. Sporn, M. Y. Liu, and J. J. Fredberg. Cellular biomechanics in the lung. Am. J. Physiol. Lung Cellular Mol. Physiol. 283(3):L503–L509, 2002.
Weiss, L., G. Elkin, and E. Barberaguillem. The differential resistance of B16 wild-type and F10 cells to mechanical trauma in vitro. Invasion Metastasis 13(2):92–101, 1993.
Wood, S. T., Computational approaches to understand phenotypic structure and constitutive mechanics relationships of single cells. Bioengineering, 2011.
Xavier, J. B., A. Schnell, S. Wuertz, R. Palmer, D. C. White, and J. S. Almeida. Objective threshold selection procedure (OTS) for segmentation of scanning laser confocal microscope images. J. Microbiol. Methods 47(2):169–180, 2001.
Acknowledgments
The authors would like to acknowledge the technical support provided by Shekhar Kanetkar and Zhong Qin of MSC Software and the financial support provided by the following grants from the National Institutes of Health: K25 HL 092228 and P20 RR-016461, and the National Science Foundation: CCF-0845593 and EPS-0903795.
Conflicts of interest
No conflict of interest apply to this manuscript and related work.
Author information
Authors and Affiliations
Corresponding author
Additional information
Associate Editor Cheng Dong oversaw the review of this article.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wood, S.T., Dean, B.C. & Dean, D. A Computational Approach to Understand Phenotypic Structure and Constitutive Mechanics Relationships of Single Cells. Ann Biomed Eng 41, 630–644 (2013). https://doi.org/10.1007/s10439-012-0690-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10439-012-0690-5