Abstract
Objectives
To characterize a novel ene-reductase from Meyerozyma guilliermondii and achieve the ene-reductase-mediated reduction of activated C=C bonds.
Results
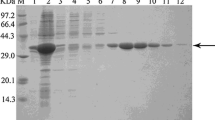
The gene encoding an ene-reductase was cloned from M. guilliermondii. Sequence homology analysis showed that MgER shared the maximal amino acid sequence identity of 57 % with OYE2.6 from Scheffersomyces stipitis. MgER showed the highest specific activity at 30 °C and pH 7 (100 mM sodium phosphate buffer), and excellent stereoselectivities were achieved for the reduction of (R)-carvone and ketoisophorone. Under the reaction conditions (30 °C and pH 7.0), 150 mM (R)-carvone could be completely converted to (2R,5R)-dihydrocarvone within 22 h employing purified MgER as catalyst, resulting in a yield of 98.9 % and an optical purity of >99 % d.e.
Conclusion
MgER was characterized as a novel ene-reductase from yeast and showed great potential for the asymmetric reduction of activated C=C bonds of α,β-unsaturated compounds.



Similar content being viewed by others
References
Bougioukou DJ, Walton AZ, Stewart JD (2010) Towards preparative-scale, biocatalytic alkene reductions. Chem Commun 46:8558–8560
Chaparro-Riggers JF, Rogers TA, Vazquez-Figueroa E, Polizzi KM, Bommarius AS (2007) Comparison of three enoate reductases and their potential use for biotransformations. Adv Synth Catal 349:1521–1531
Dong Y, McCullough KJ, Wittlin S, Chollet J, Vennerstrom JL (2010) The structure and antimalarial activity of dispiro-1,2,4,5-tetraoxanes derived from (+)-dihydrocarvone. Bioorg Med Chem Lett 20:6359–6361
Durchschein K, Hall M, Faber K (2013) Unusual reactions mediated by FMN-dependent ene- and nitro-reductases. Green Chem 15:1764–1772
Fu Y, Hoelsch K, Weuster-Botz D (2012) A novel ene-reductase from Synechococcus sp. PCC 7942 for the asymmetric reduction of alkenes. Proc Biochem 47:1988–1997
Fu Y, Castiglione K, Weuster-Botz D (2013) Comparative characterization of novel ene-reductases from Cyanobacteria. Biotechnol Bioeng 110:1293–1301
Gao X, Ren J, Wu Q, Zhu D (2012) Biochemical characterization and substrate profiling of a new NADH-dependent enoate reductase from Lactobacillus casei. Enzym Microb Technol 51:26–34
Hall M, Stueckler C, Ehammer H, Pointner E, Oberdorfer G, Gruber K, Hauer B, Stuermer R, Kroutil W, Macheroux P, Faber K (2008) Asymmetric bioreduction of C=C bonds using enoate reductases OPR1, OPR3 and YqjM: enzyme-based stereocontrol. Adv Synth Catal 350:411–418
Kataoka M, Kotaka A, Thiwthong R, Wada M, Nakamori S, Shimizu S (2004) Cloning and overexpression of the old yellow enzyme gene of Candida macedoniensis, and its application to the production of a chiral compound. J Biotechnol 114:1–9
Lowe JR, Tolman WB, Hillmyer MA (2009) Oxidized dihydrocarvone as a renewable multifunctional monomer for the synthesis of shape memory polyesters. Biomacromolecules 10:2003–2008
Mueller NJ, Stueckler C, Hauer B, Baudendistel N, Housden H, Bruce NC, Faber K (2010) The substrate spectra of pentaerythritol tetranitrate reductase, morphinone reductase, N-ethylmaleimide reductase and estrogen-binding protein in the asymmetric bioreduction of activated alkenes. Adv Synth Catal 352:387–394
Ni Y, Yu HL, Lin GQ, Xu JH (2014) An ene reductase from Clavispora lusitaniae for asymmetric reduction of activated alkenes. Enzym Microb Technol 56:40–45
Patterson-Orazem A, Sullivan B, Stewart JD (2014) Pichia stipitis OYE 2.6 variants with improved catalytic efficiencies from site-saturation mutagenesis libraries. Bioorg Med Chem 22:5628–5632
Paul CE, Gargiulo S, Opperman DJ, Lavandera I, Gotor-Fernández V, Gotor V, Taglieber A, Arends IWCE, Hollmann F (2013) Mimicking nature: synthetic nicotinamide cofactors for C=C bioreduction using enoate reductases. Org Lett 15:180–183
Pei XQ, Xu MY, Wu ZL (2016) Two “classical” old yellow enzymes from Chryseobacterium sp. CA49: broad substrate specificity of Chr-OYE1 and limited activity of Chr-OYE2. J Mol Catal B Enzym 123:91–99
Pompeu YA, Sullivan B, Walton AZ, Stewart JD (2012) Structural and catalytic characterization of Pichia stipitis OYE 2.6, a useful biocatalyst for asymmetric alkene reductions. Adv Synth Catal 354:1949–1960
Selvam P, Sonavane SU, Mohapatra SK, Jayaram RV (2004) Selective reduction of alkenes, α,β-unsaturated carbonyl compounds, nitroarenes, nitroso compounds,N,N-hydrogenolysis of azo and hydrazo functions as well as simultaneous hydrodehalogenation and reduction of substituted aryl halides over PdMCM-41 catalyst under transfer hydrogen conditions. Tetrahedron Lett 45:3071–3075
Tuttle Jamison B, Ouellet Stephane G, MacMillan DWC (2006) Organocatalytic transfer hydrogenation of cyclic enones. J Amer Chem Soc 128:12662–12663
Yin B, Deng J, Lim L, Yuan YA, Wei D (2015) Structural insights into stereospecific reduction of α,β-unsaturated carbonyl substrates by old yellow enzyme from Gluconobacter oxydans. Biosci Biotechnol Biochem 79:1–12
Acknowledgments
This work was financially supported by the National Key Basic Research Development Program of China (“973” Program, No. 2012CB721003), the Natural Science Foundation of China (No. 21276084), Shanghai Natural Science Foundation (No. 15ZR1408600) and National Major Science and Technology Projects of China (No. 2012ZX09304009).
Supporting information
Supplementary Figure 1—Multiple sequence alignment of MgER with the other nine Old Yellow Enzymes. Substrate binding sites are indicated.
Supplementary Figure 2—Phylogenetic relationship of amino acid sequences of MgER to other Old Yellow Enyzmes with known function.
Supplementary Figure 3—GC-MS spectrum of (2R,5R)-dihydrocarvone prepared by MgER. GC-EI-MS m/z (M+ 152 for C10H16O) 137, 109, 95, 81, 67, 55.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the Electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhang, B., Zheng, L., Lin, J. et al. Characterization of an ene-reductase from Meyerozyma guilliermondii for asymmetric bioreduction of α,β-unsaturated compounds. Biotechnol Lett 38, 1527–1534 (2016). https://doi.org/10.1007/s10529-016-2124-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10529-016-2124-1