Abstract
Species distribution models are widely used in conservation planning, but obtaining the necessary occurrence data can be challenging, particularly for rare species. In these cases, citizen science may provide insight into species distributions. To understand the distribution of the newly described and Critically Endangered Amazona lilacina, we collated species observations and reliable eBird records from 2010–2020. We combined these with environmental predictors and either randomly generated background points or absence points generated from eBird checklists, to build distribution models using MaxEnt. We also conducted interviews with people local to the species’ range to gather community-sourced occurrence data. We grouped these data according to perceived expertise of the observer, based on the ability to identify A. lilacina and its distinguishing features, knowledge of its ecology, overall awareness of parrot biodiversity, and the observation type. We evaluated all models using AUC and Tjur R2. Field data models built using background points performed better than those using eBird absence points (AUC = 0.80 ± 0.02, Tjur R2 = 0.46 ± 0.01 compared to AUC = 0.78 ± 0.03, Tjur R2 = 0.43 ± 0.21). The best performing community data model used presence records from people who were able recognise a photograph of A. lilacina and correctly describe its distinguishing physical or behavioural characteristics (AUC = 0.84 ± 0.05, Tjur R2 = 0.51± 0.01). There was up to 92% overlap between the field data and community data models, which when combined, predicted 17,772 km2 of suitable habitat. Use of community knowledge offers a cost-efficient method to obtain data for species distribution modelling; we offer recommendations on how to assess its performance and present a final map of potential distribution for A. lilacina.
Similar content being viewed by others
Introduction
Understanding species distributions is essential for conservation planning (Wilson et al. 2005) but for species that are rare, sparsely distributed, or inconspicuous, this information is often lacking. In such cases, species distribution models (SDMs) and their outputs, can be particularly useful, as long as they are based on ecological theory and built using accurate data (Guisan and Thuiller 2005). SDMs allow the probability of occurrence to be predicted in un-surveyed areas, which can inform future field investigations and have many important conservation applications (e.g. Pearce and Lindenmayer 1998; Araújo et al. 2004). For all SDMs, species presence data are needed. Traditionally this comes from direct species observations or museum records, but more recently scientists have looked to integrate different sources of data, such as citizen science, to make better inferences of the true distribution of species (Amano et al. 2016; Coxen et al. 2017; Fletcher et al. 2019; Steen et al. 2019; Isaac et al. 2020).
The quality of outputs gained from SDMs is affected by factors such as data type, sampling bias and imperfect detection (Lahoz-Monfort et al. 2014; Guillera-Arroita et al. 2015). MaxEnt is one of the most commonly used methods for deriving SDMs and has been shown to produce useful models even when dealing with small sample sizes (Wisz et al. 2008; Elia et al. 2015). Whilst other methods require absence data to be collected, MaxEnt uses presence data combined with a background sample drawn randomly from the study area (Phillips et al. 2006, Phillips and Dudík 2008; Elith et al. 2011). Both presence-absence and presence-background data methods have limitations; namely that presence data often do not represent an unbiased sample of locations at which the species is present, and that absence data can lead to the inclusion of false absences (Guillera-Arroita et al. 2015). These limitations must be considered against the proposed use of model outputs; for instance, presence-background data may be sufficient when outputs are to be used to direct further field investigations, but insufficient if outputs are to directly inform land management for conservation (Lahoz-Monfort et al. 2014). The predictive ability of models may also be reduced if imperfect detection is not accounted for, and may result in outputs being more likely to predict areas in which the species is easier to observe, rather than where it is more likely to occur. It is therefore essential that the effects of imperfect detection are minimised by ensuring a sufficiently large sampling effort at surveyed locations (Lahoz-Monfort et al. 2014).
For species where field observations are lacking, citizen science data is a valuable and widely used resource (Brook and McLachlan 2008) which can help determine species presence, absence or abundance (Melovski et al. 2018; Díaz-Ruiz et al. 2019; Ghoshal et al. 2019; Skroblin et al. 2021). Some methods allow large volumes of data to be collected more cost effectively than traditional field survey methods, for example postal surveys (FitzGibbon & Jones 2006), telephone interviews (Mallory et al. 2003) and social media (Pace et al. 2019). Often this information is used to supplement ‘expert’ data by guiding further field surveys (Hart & Upoki 1997; O’Brien et al 1998; Chaiyes et al. 2017) but in some cases it is shown to be just as accurate as the equivalent ‘expert’ data, providing that some form of filter for reliability is incorporated (Polfus et al. 2014). Recently, a number of studies have even shown that georeferenced occurrence data collected through citizen science platforms and online biodiversity databases such as eBird, can be used to build accurate SDMs (Bradsworth et al. 2017; Coxen et al. 2017; Fournier et al. 2017; Saunders et al. 2020). However, it is important to note that all opportunistically collected citizen science data present additional challenges such as spatial biases and variation in observer skill (Isaac and Pocock 2015; Johnston et al. 2020) and online recording schemes such as eBird create barriers by requiring observations to be collected and submitted in a particular way.
Within all types of citizen science data, there is variation in accuracy. For example studies have shown that ‘freelisting’ (Bernard 2006), a quick survey method where participants are asked to list the species they see in their local area, can result in people reporting species that do not occur and omitting ones that do (Can and Togan 2009; Díaz-Ruiz et al. 2019). However, the cost efficiency of citizen science may compensate for reduced accuracy depending the data collected and extent of errors (Gardiner et al. 2012). If citizen science data are to be used to infer information about distribution, and as input data for the creation of SDMs, some method of boosting data accuracy or accounting for level of expertise is essential (Kosmala et al. 2016; Johnston et al. 2019). Previous studies have used prior selection of participants i.e. only interviewing key informants selected by community leaders due to their perceived expertise (Mallory et al. 2003; Lopes et al. 2018). Others have developed some kind of scoring system, to determine data accuracy (Frey et al. 2013) by only regarding contributions from participants who are able to recognise photographs of the study species and provide accurate location information (Ghoshal et al. 2019), or by using photographs of non-native species to assess participants identification skills (O’Brien et al. 1998).
To further our understanding of the distribution of a newly described and Critically Endangered parrot species Amazona lilacina (Biddle et al. 2020; BirdLife International. 2020), we:
-
1.
Built distribution models using all known locality records of A. lilacina from our own observations, those from expert ornithologists, and reliable eBird records (2010–2020);
-
2.
Collected data on local peoples’ experiences and observations of wild A. lilacina through structured face-to-face interviews;
-
3.
Grouped community interview data based on different quality filters and used these data to build distribution models;
-
4.
Determined the best performing distribution models built from species records and community reports, and compared their outputs in order to direct future field investigation.
Methods
Study area
Amazona lilacina, a species recently split from the A. autumnalis group, is found in the coastal region of Ecuador where its small population is sparsely distributed around dry forests and mangrove ecosystems (Biddle et al 2020). These habitats are described as amongst the most imperilled ecosystems on earth (Dodson and Gentry 1991). During the day-time A. lilacina is highly inconspicuous, feeding silently in the forest canopy in small groups which presents difficulty in using traditional field survey methods to collect presence data (Ridgely and Greenfield 2001a). However, in the evenings birds will form conspicuous groups and fly to communal roost sites (Berg and Angel 2006) which means that communities living anywhere on this flight path, are often aware of the species presence.
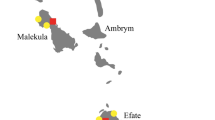
The rural coastal communities are considered to be in the most deprived areas of Ecuador, with almost one quarter of all people living in multidimensional poverty (Mideros 2012). The deprivation gap regarding food and water, education, communication, and housing, is greater here than in any other part of the country (Mideros 2012). Within our sampled communities (Fig. 1a), people mainly make a living as farmers, fishers or crab fishers, and 60% have either none, or only primary level schooling. Many communities in this region are highly inaccessible, especially in the rainy season and 57% of people we surveyed had lived in their village their entire lives. The flow of information into and out of these communities is reported to be infrequent, with only 40% of households having access to one form of telecommunication (radio, television, phone, computer) (Mideros 2012).
a Locations of all households taking part in interviews, all records of Amazona lilacina collated between 2010 – 2020 and, b eBird absence points, representing all complete checklists that did not report A. lilacina, and random background points matching the number of eBird absence points available, within a 30 km buffer of all A. lilacina presence records
Field observations and eBird records
Observational data were collected during ten field trips led by RB, lasting two to three weeks each (November 2012, January and August 2014, November 2015, August 2016, January and March 2017, February 2018, January and August 2019). Data collection was informed by: (1) existing information on known distribution and habitat use (Juniper and Parr 1998; Ridgely and Greenfield 2001a, b; Berg and Angel 2006; Forshaw and Knight 2010; Athanas and Greenfield 2016); (2) information on habitat distribution from Google Earth and the Ministerio del Ambiente ecosystem map; (3) direct communication with local NGOs, ornithologists, local guides and bird tour companies. All sightings of perched A. lilacina made by RB, ISP, MP, Fundación Pro-Bosque staff, Fundación Jambeli staff, and Juan Freile between 2010 and 2020 were georeferenced (sightings of birds in flight were omitted).
All eBird data for Ecuador, including observations and sampling data were downloaded in December 2020. To ensure that no records were missed due to changing taxonomic nomenclature, data were filtered to include all birds recorded as A. autumnalis (which included A. a. lilacina and A. a. salvini) between 01/01/2010 and 31/12/2020. Records that were not deemed as A. lilacina based on either photographic evidence or location (i.e. within the Esmeraldas province) were removed, as were records that were already represented by our own observations (within 1 km). To avoid misrepresentation of location, all records that were reported as “general area” which implies the record does not correspond to that exact location were removed, as were records with survey effort > 5 h and > 5 km in length (Johnston et al 2019). Finally, locations of parrots within urban locations in the big city of Guayaquil (visualised on Google Earth) were removed to avoid escaped pets or captive birds being included in models.
Distribution models from field observations and eBird records
The MaxEnt function of the package ‘dismo’ (Hijmans et al. 2020) in R (version 3.6.0, R Core Team 2019) was used to create species distribution models from field observations and eBird records, referred to from now on as the field models. These were first built using eBird absence points generated by filtering for all complete checklists within our study area that did not report the presence of A. autumnalis (A. a. salvini or A. a. lilacina) (Fig. 1b). Absence points were also limited to checklists that were < 5 km in length, < 5 h in duration and with fewer than ten observers (Johnston et al. 2019), and to a buffer of 30 km from all field observations and eBird records. Our second and third field models were built using random background points generated in ArcGIS (Version 10.8.1) from within the same buffer: the second model had 4597 and the third had the same number as eBird absences available (458). Spatial autocorrelation was controlled for by limiting points to one per 1 km using the R package ‘spThin’ (Aiello-Lammens et al. 2015). A set of interpolated bioclimatic predictor variables available from WorldClim (https://www.worldclim.com/bioclim) representing different measures of temperature and rainfall, plus additional predictors thought to have some biological significance for the species were used: Normalised Difference Vegetation Index (NDVI) from the monthly MODIS product over the period 2010–2015 as a proxy of vegetation cover; distance to mangrove (Hamilton and Casey 2016) and distance to the nearest river (Military Geographic Institute, IGM). Predictors were checked for pairwise correlation across random points within the study area, using pair plots (Zuur et al. 2010); where correlation coefficients between pairs of predictors were ≥ 0.70, the less biologically meaningful predictor was removed. The final variables were; distance to the mangrove, distance to a river, annual mean NDVI and NDVI seasonality, mean diurnal temperature range, annual mean temperature and temperature seasonality, precipitation of wettest month, precipitation of coldest quarter and precipitation of driest month. To allow comparison between the field and community models, we averaged predictor values across 9 km2 at all points used in all models to reflect respondents’ reference to their ‘local area’, which could encompass areas of community owned land > 1 km away from their house. To ensure this did not affect model outputs or accuracy we trialled models built using predictor values at the exact location, compared to those averaged over 9 km2, and found no difference.
Models were evaluated with AUC and Tjur R2 (Tjur 2009) over five-fold cross validation; the mean evaluation metrics and their standard deviation are presented. AUC measures how well model predictions discriminate between presence and absence (Wisz et al. 2008). Tjur R2 represents the difference between the mean model value at the presence locations and the mean value at the absence / background locations. All the data were included in the final models. Finally, we present variable importance scores, with permutation values > 10%, with a high value indicating that the model depends heavily on that variable (Phillips et al. 2006) and response plots for the most accurate field model.
Community questionnaires and response filtering
Researcher–led questionnaires were carried out to identify areas that were reported by local people to be occupied by A. lilacina. Communities were chosen to be included in this study due to their close proximity to dry lowland forests (within approximately 10 km), identified using the Ministerio del Ambiente ecosystem map. Furthermore, all communities surveyed were inside or within 70 km of the species Extent of Occurrence (Biddle et al 2020). A pilot study was conducted after which interviews were carried out in January-July 2017. Questionnaires were conducted in Spanish by a local Ecuadorian researcher (ISP), with only the interviewer and respondent present (Tourangeau and Yan 2007). We aimed to survey a minimum of three households per community representing a cross section of demographic groups, but often this depended on the availability of participants and the size of the community. In all cases, prior verbal consent was obtained, and although less than fifteen people did not complete interviews, interviewees could decline from contributing once the purpose of the research was explained (Online Resource 1).
The location of each questionnaire, normally by the participant’s house, was recorded and participants were asked to respond with reference to their immediate local area which included their house, garden, and local community land. Demographic information regarding age, gender, level of schooling, and how long they had lived in the village, was collected, but interviews were anonymous, and data were coded to ensure that no individuals could be identified. Interviewees were not made aware of the species in concern before starting the interview, during which they were asked to name and describe which parrot species (if any) they see in their local area, then confirm from a selection of ten parrot photographs (the order of which was rotated at random between surveys) (Table 1). If a participant confirmed they currently (within the last year) see A. lilacina at their location, they were then asked a number of questions designed to help assess the accuracy of this information. Each interview (Online Resource 2) took approximately 20 min to complete.
To examine the influence of accuracy of community data, we filtered responses according to the ability to recognise the species, knowledge of its distinguishing features, overall awareness of parrot biodiversity, and observation type (i.e., if the bird was seen flying, nesting, perched or feeding). We created six groups of responses to represent realistic scenarios that may be used to select which observations to include in distribution investigations (Table 2). We created a further 11 groups which represented all possible combinations of groups three-six, for example group seven represented a group of participants who had answered correctly for all of groups three, four, five and six (Online Resource 3).
Distribution models from community data
We created distribution models based on groups of community data with varying levels of accuracy as listed in Table 2; the community models. Each participant’s response was associated with a location representing a 1 km2 pixel on our distribution maps. These presence locations were combined with environmental variables and background points following the same methods as for the field model. All background points were restricted to buffers of 30 km from community survey presence points. We averaged predictor values across the 9 km2, as for the field model, to reflect respondents’ reference to their ‘local area’, which could encompass additional areas of community owned land. In order to evaluate the accuracy of the community data models, we use the same methods as for the field models; AUC and Tjur R2 (Tjur 2009) over five-fold cross validation. We present these, alongside permutation values where their contribution to the model is > 10% for all models, and the habitat suitability output and response plots for the best performing model.
Model comparison
Once we had identified the best performing field observation model and community data model, we compared the overlap between their habitat suitability outputs. These outputs are interpreted as maps of potential distribution with values indicating the level of habitat suitability for each pixel, on a scale of zero to one. There are several methods used to compare model outputs (Galante et al. 2018). We chose Moran’s I which represents the difference between suitability values at each cell, and the relative rank coefficient which estimates the probability that the relative suitability ranking for a patch of habitat cells is the same for the two models (Warren and Seifert 2011). We calculated these using the niche overlap function in ENMTools (Warren et al. 2010). Both methods produce metrics which range from zero (no overlap) to one (complete overlap).
To predict areas of potential distribution, it was necessary to classify areas as either ‘suitable’ or ‘unsuitable’ depending on their model value. Many thresholding rules are justified for presence-only occurrence data (Peterson et al. 2011). We chose the 10% omission rate threshold (Galante et al. 2018) where the model value which includes 90% of the values predicted at the presence locations used to create that model, is applied as a threshold to the habitat suitability output to distinguish between presence and absence. We calculated and applied this independently to the two best performing models. We present a final combined map of distribution that represents areas predicted as suitable or not by either of the final models. We extracted the values for the top three predictor variables from the best performing models, in areas where both models predicted presence, compared to areas where only the field model or only the community model did, and plotted these using the R package ‘ggplot2′ (Wickham 2016).
Predictors of community data performance
Once the best performing community data model been determined, a generalized linear mixed model (GLMM) was conducted in R (version 3.6.0, R Core Team, 2019) using the package ‘lme4′ (Bates et al. 2020). The binomial response of whether or not a participant was included in the response group used to build that model was analysed to determine any effects of participants’ social demographics: gender, level of schooling, age and number of years in the village. Only communities where at least one wild A. lilacina observation had been reported were included, and the community location was included as a random effect. We checked for correlation between the age and number of years spent in the village using Pearson's product-moment correlation, and between gender and level of schooling (some or none) using a Chi-squared test of independence, and only included non-correlated variables in our GLMM.
Results
Field observations and distribution model
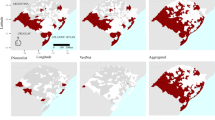
Our field observations generated a total of 132 occurrence points. A further 14 locations from eBird were included, to create a final dataset of 146 A. lilacina presence locations. These were reduced to 59 (47 field observations and 12 eBird records) during the spatial rarefication process, combined with either: 458 eBird absence points (model 1); 4597 randomly generated background points (model 2) or; 458 randomly generated background points (model 3) and entered into model building with the ten non-correlated predictor variables. The resulting mean of five-fold cross validation AUCs were 0.78 ± 0.03, 0.80 ± 0.02, 0.79 ± 0.02 and the resulting mean of five-fold cross validation Tjur R2s were 0.43 ± 0.21, 0.46 ± 0.01 and 0.41 ± 0.01 for models 1 to 3, respectively. Therefore, field model 2 was considered to be the best performing model (Table 3). The habitat suitability output from model 2 shows that the suitable habitat follows the Chongón Colonche mountain range, from Guayaquil north-west towards the coast, with additional suitable areas in the far south of the country bordering Peru, and the north of the study area in mid-Manabí (Fig. 2a). Environmental variables that showed a permutation importance of > 10% were annual mean NDVI, distance to the mangrove, and temperature seasonality and response plots (Fig. 2b) suggest that suitability of habitat is associated with close distance to mangrove and a relatively high annual mean NDVI.
Community questionnaires and reliability scoring
A total of 404 people from 72 communities took part in questionnaires, including 183 women and 221 men, with an average of 5.6 households per community (min 2, max 23). There was a variety of schooling levels, from none (31), primary (214), secondary (128), to university (31) and in how long participants had lived in their community (1–84 years) but the majority (88%) had lived there for ten or more years. Of the 404 participants, 393 reported seeing parrots in general. Although it was posed in our questionnaires that participants should answer with reference to birds seen in the wild, when asked “where did you see this bird?” 15 respondents replied “as a pet” - these 15 responses were removed from the community models.
Distribution models from community data
After filtering community data based on the six groups in Table 1, and creating combination groups where participants answered positively for multiple categories, each group had a sample size of ≥27 (27–155). After spatial thinning all datasets contained ≥18 (18–67) georeferenced occurrence points. Each group of points was combined with 3,931 background points and the same ten non-correlated predictor variables as those included in the field models. Models were built based on groups one to six of data, and then all 11 possible combinations of groups three to six. None of the combination models improved the performance of the model (Online Resource 3). The mean of five-fold cross validation AUC for the six main models was > 0.74 ± 0.03 and Tjur R2 > 0.39 ± 0.02. Based on these values, model 3 is the best performing community model (Table 4). The habitat suitability map of community model 3 shows a similar area of suitable habitat to the field data model, but with additional increased suitability predicted along the coastline (Fig. 3a). Environmental variables with a permutation importance of > 10% were distance to mangrove and temperature seasonality, and response plots for this model suggest that suitability of habitat is associated with areas closer to mangroves (Fig. 3b).
a The habitat suitability output from the best performing community data model, built using 53 reports where participants were able to recognise a photograph of the species and provide one or more physical or behavioural characteristics specific to A. lilacina. b The variable response plots for this model
Model comparison
After calculating and applying thresholds to the best performing field and community models, the field model predicts 13,969 km2 of suitable habitat and the community model predicts 13,067 km2 (Table 5). When we combine these threshold habitat suitability outputs, they overlap in 9314 km2 of predicted suitable habitat, the community data model predicts a further 3753 km2 that the field data does not, and the field data model predicts a further 4655 km2 that the community model does not (Fig. 4). The top three predictor variables from both of these models were; distance to mangrove, temperature seasonality and mean annual NDVI. When plotting the values from predicted presence areas by both models, just the field model or just the community model, areas that are predicted by only the community model have a slightly lower mean annual NDVI and are closer to mangroves than areas only predicted by the field model (Fig. 5). There is a high level of overlap between the field data and community data habitat suitability outputs (before applying a threshold). The relative rank coefficient, which estimates the probability that the relative suitability ranking for a patch of habitat cells is the same for the two models, is 0.82, and the Moran’s I, which represents the difference between suitability values at each cell, is 0.92 (Table 5).
After calculating and applying thresholds independently to the two best performing models, their predicted suitable habitat overlaps in 9314 km2, but the community data model predicts a further 3753 km2 that is suitable, and the field data model predicts a further 4655 km2 that is suitable for A. lilacina
Box plots showing predictor values in areas predicted as suitable (after applying a threshold) by both the best performing community and field data models, only the field data model, and only the community data model. The predictors with a permutation importance of > 10% in the final models were included; mean annual NDVI a distance to mangrove b and temperature seasonality c
Predictors of community data performance
Of the 52 communities where at least one observation of wild A. lilacina was made, and thus species presence was likely, 35% (105/304) of participants were included in community data group with the best model performance. These 105 participants (70 men and 35 women) were able to either name or recognise a photo of the species, and describe one of its distinguishing physical or behavioural characteristics (Table 6). There was a high correlation coefficient of 0.70 (p < 0.001) between the number of years lived in the village and the age of a participant. Additionally, gender and level of schooling were significantly correlated (X2 = 8.24, df = 1, p = 0.004). Therefore, we only included the number of years a participant had lived in the village, and the participant’s gender in our GLMM. This revealed that of participants living in areas where A. lilacina was likely to be present, men were more likely to be included in the better performing community data group than women (Coefficient value: 0.62 ± 0.31, p = 0.04), which is likely due to their spending more time outdoors in traditionally male working roles. The number of years a participant had lived in the community (Coefficient value: 0.012 ± 0.007, p = 0.14) had no significant effect.
Discussion
We found that both field data and citizen science data in the form of community surveys were able to produce accurate species distribution models and their outputs had an overlap of 92%. When using field data, we found that models built using background points performed better than those built using absence points generated by eBird checklists, possibly due to the low frequency of eBird records in our study area. When using community data, we found the best performing models were those built using reports from observers who could name or recognise a photograph of A. lilacina and correctly describe at least one distinguishing physical or behavioural characteristic.
Recent studies have shown that web-based citizen science projects and online biodiversity databases can be used to build reliable species distribution models (e.g. Saunders et al. 2020; Langham et al. 2015; Fournier et al. 2017). This study presents evidence that in areas where there are substantial barriers to web-based citizen science projects, for example in socio-economically deprived areas (e.g. Hobbs and White 2012), community surveys can overcome these barriers and produce accurate species distribution models. This is of particular use for newly described and rare species. Gender disassociation in local ecological knowledge is not uncommon (Kai et al. 2014; Aswani et al. 2018); we found that men were more likely to provide accurate answers than women and suggest that this is due to a gender difference in traditional working roles (Voeks 2007; Ayantunde et al. 2008) which allows men to spend more time outdoors. Erosion of local ecological knowledge is a global trend (Aswani et al. 2018) and we support the continuation of community wide engagement projects to minimise this risk, with a focus on support for women to enable them to engage with conservation.
After applying thresholds to our best performing field and community data models, they overlapped in their predictions of suitable habitat by 92% (in 9314 km2). The level of overlap we see between our community and field data models is greater than seen in similar comparison of eBird community data and field-based satellite tracking data of Band-tailed Pigeons Patagioenas fasciata (Coxen et al. 2017). Our community data model predicts a further 3753 km2 of suitable habitat that our field data model does not. These areas were closer to mangroves than areas predicted only by the field data model. This may be due to a factor of species detectability; A. lilacina are more detectable (highly vocal) when flying over to mangrove communal roost sites, so perhaps more likely to be seen by local communities in this habitat compared to when they are foraging inconspicuously in the dry forest (Ridgely and Greenfield 2001a). It is also possible that these areas represent locations in which local people have memories of the species occurring in the past, in which they no longer occur and thus were not recorded during field surveys. Our field data model predicts a further 4655 km2 of suitable habitat that our community data models do not, and in areas with a slightly higher mean annual NDVI than areas predicted only by the community model.
Similarly to Frey et al. (2013), we found variation in the accuracy of community data models built using different methods to filter interview responses. Our best performing model used a filter whereby participants needed to recognise a photograph of the species and provide a reliable description of how they distinguish it from other parrot species in their area. This suggests that, particularly in areas where many similar taxa may occur, the key to assessing the accuracy of information may be simply to ensure that participants are referring to the correct species. This draws parallels with checks that are in place for citizen science online databases such as eBird where records are flagged for systematic review and confirmed by a regional expert prior to their acceptance (Sullivan et al. 2014). It also supports the work of Frey et al. (2013) who conclude that, for easily-identifiable species at least, distribution modelling is possible using anecdotal reports. Our second best community data model (1) greatly underestimated the predicted area of suitable habitat. This group was based on the ‘freelisting’ method, where participants needed to name the parrot species in their area without any prior information or prompting. Previous studies using the freelisting method have yielded questionable results (e.g. Can and Togan 2009; Díaz-Ruiz et al. 2019) and we believe in our case, it was due to a very small sample size of participants who had the required natural history expertise to name this rare parrot species without any prompting or information.
We found that using identification of other parrot species, to measure overall biodiversity knowledge and therefore accuracy of answers, did not produce the most accurate results. This may be due to A. lilacina’s unique daily migration behaviour, in some cases flying directly over villages and becoming conspicuous to many community members, not just those that are skilled at identifying multiple parrot species. Alternatively, it is possible that the two parrot species whose identification we assessed as a measure of reliability are incorrectly believed to be common and widespread throughout our study area (Ridgely and Greenfield 2001b; Freile and Restall 2018). Identification of other closely related species was not a good measure of data quality either in surveys investigating the distribution of a native pheasant species – results showed frequent misidentification of an ‘imposter’ pheasant photograph, but reliable information about the native pheasant was still generated (O’Brien et al. 1998).
Our distribution models based on field data and high quality community knowledge represent the first of their kind for the newly described and Critically Endangered A. lilacina, and have important conservation implications. With an estimated population size of just ~ 1,000 birds, and a suggested recent 60% population decline in parts of the range (Biddle et al. 2020), our results have identified new areas to survey. It is important to note that our model predictors did not include factors such as poaching that may have a strong impact on occupancy (Robinson et al. 2010). Whilst conducting community surveys for this study, we discovered a new large roost, unknown previously to local and international ornithologists, located near a socio-economically deprived coastal community, on a mangrove island. Even local residents, because of the conflict with pirates, deem this area as unsafe. We therefore recommend that when parts of a species range fall within areas that are rarely visited by outsiders, the combined knowledge of communities local to that species is likely to be much greater than that of external scientists or researchers, and should thus be used to enhance and supplement traditional field survey methods.
References
Aiello-Lammens ME, Boria RA, Radosavljevic A, Vilela B, Anderson RP (2015) spThin: An R package for spatial thinning of species occurrence records for use in ecological niche models. Ecography 38:541–545
Amano T, Lamming JDL, Sutherland WJ (2016) Spatial Gaps in Global Biodiversity Information and the Role of Citizen Science. Bioscience 66:393–400
Araújo MB, Cabeza M, Thuiller W, Hannah L, Williams PH (2004) Would climate change drive species out of reserves? An assessment of existing reserve-selection methods. Glob Change Biol 10:1618–1626
Aswani S, Lemahieu A, Sauer WHH (2018) Global trends of local ecological knowledge and future implications. PLoS ONE 13:1–19
Athanas N, Greenfield PJ (2016) Birds of Western Ecuador: A Photographic Guide. Princeton University Press, USA
Ayantunde AA, Briejer M, Hiernaux P, Udo HMJ, Tabo R (2008) Botanical knowledge and its differentiation by age, gender and ethnicity in Southwestern Niger. Hum Ecol 36:881–889
Bates D et al (2020) Package ‘lme4’
Berg KS, Angel RR (2006) Seasonal roosts of Red-lored Amazons in Ecuador provide information about population size and structure. J Field Ornithol 77:95–103
Bernard RH (2006) Research methods in anthropology; qualitative and quantitative approaches. Altamira Press, USA
Biddle R, Solis-Ponce I, Cun P, Tollington S, Jones M, Marsden S, Devenish C, Horstman E, Berg K, Pilgrim M (2020) Conservation status of the recently described Ecuadorian Amazon parrot Amazona lilacina. Bird Conserv Int 30(4):586–598
BirdLife International (2020) Amazona lilacina. The IUCN Red List of Threatened Species 2020: e.T22728296A181432250. https://doi.org/10.2305/IUCN.UK.2020-3.RLTS.T22728296A181432250.en
Bradsworth N, White JG, Isaac B, Cooke R (2017) Species distribution models derived from citizen science data predict the fine scale movements of owls in an urbanizing landscape. Biol Conserv 213:27–35. https://doi.org/10.1016/j.biocon.2017.06.039
Brook RK, McLachlan SM (2008) Trends and prospects for local knowledge in ecological and conservation research and monitoring. Biodivers Conserv 17:3501–3512
Can ÖE, Togan I (2009) Camera trapping of large mammals in Yenice Forest, Turkey: local information versus camera traps. Oryx 43:427–430
Chaiyes A, Duengkae P, Wacharapluesadee S, Pongpattananurak N, Olival KJ, Hemachudha T (2017) Assessing the distribution, roosting site characteristics, and population of Pteropus lylei in Thailand. Raffles Bull Zool 65:670–680
Coxen CL, Frey JK, Carleton SA, Collins DP (2017) Species distribution models for a migratory bird based on citizen science and satellite tracking data. Global Ecol Conserv 11:298–311. https://doi.org/10.1016/j.gecco.2017.08.001
Díaz-Ruiz F, Caro J, Ferreras P, Delibes-Mateos M (2019) Assessing mammal community composition in the Huinay Biological Reserve (Chile) through questionnaire surveys: biases associated with respondents. Galemys, Spanish Journal of Mammalogy 31:1–9
Dodson CH, Gentry AH (1991) Biological Extinction in Western Ecuador. Ann Mo Bot Gard 78:273
Elia JD, Haig SM, Johnson M, Marcot BG, Young R (2015) Activity-specific ecological niche models for planning reintroductions of California condors (Gymnogyps californianus). Biol Conserv 184:90–99. https://doi.org/10.1016/j.biocon.2015.01.002
Elith J, Phillips S, Hastie T, Dudík M, Chee Y, Yates C (2011) A statistical explanation of MaxEnt for ecologists. Divers Distrib 17(1):43–57
FitzGibbon SI, Jones DN (2006) A community-based wildlife survey: The knowledge and attitudes of residents of suburban Brisbane, with a focus on bandicoots. Wildl Res 33:233–241
Fletcher RJ, Hefley TJ, Robertson EP, Zuckerberg B, McCleery RA, Dorazio RM (2019) A practical guide for combining data to model species distributions. Ecology 100:1–15
Forshaw JM, Knight F (2010) Parrots of the world. Helm, UK
Fournier AMV, Drake KL, Tozer DC (2017) Using citizen science monitoring data in species distribution models to inform isotopic assignment of migratory connectivity in wetland birds. J Avian Biol 48:1556–1562
Freile JF, Restall R (2018) Birds of Ecuador. Helm, UK
Frey JK, Lewis JC, Guy RK, Stuart JN (2013) Use of Anecdotal Occurrence Data in Species Distribution Models: an example based on the white-nosed coati (Nasua narica) in the American southwest:327–348
Galante PJ, Alade B, Muscarella R, Jansa SA, Goodman SM, Anderson RP (2018) The challenge of modeling niches and distributions for data-poor species: a comprehensive approach to model complexity. Ecography 41:726–736. Blackwell Publishing Ltd. Available from http://doi.wiley.com/. https://doi.org/10.1111/ecog.02909 (accessed March 9, 2020)
Gardiner MM, Allee LL, Brown PMJ, Losey JE, Roy HE, Smyth RR (2012) Lessons from lady beetles: Accuracy of monitoring data from US and UK citizenscience programs. Front Ecol Environ 10:471–476
Ghoshal A, Bhatnagar YV, Pandav B, Sharma K, Mishra C, Raghunath R, Suryawanshi KR (2019) Assessing changes in distribution of the Endangered snow leopard Panthera uncia and its wild prey over 2 decades in the Indian Himalaya through interview-based occupancy surveys. Oryx 53:620–632
Guillera-Arroita G, Lahoz-Monfort JJ, Elith J, Gordon A, Kujala H, Lentini PE, Mccarthy MA, Tingley R, Wintle BA (2015) Is my species distribution model fit for purpose? Matching data and models to applications. Glob Ecol Biogeogr 24:276–292
Guisan A, Thuiller W (2005) Predicting species distribution: Offering more than simple habitat models. Ecol Lett 8:993–1009
Hamilton SE, Casey D (2016) Creation of a high spatio-temporal resolution global database of continuous mangrove forest cover for the 21st century (CGMFC-21). Glob Ecol Biogeogr 25:729–738
Hart JA, Upoki A (1997) Distribution and conservation status of Congo Peafowl Afropavo congensis in eastern Zaire. Bird Conserv Int 7:295–316
Hijmans ARJ, Phillips S, Leathwick J, Elith J, Hijmans MRJ (2020) Package ‘dismo’
Hobbs SJ, White PCL (2012) Motivations and barriers in relation to community participation in biodiversity recording. J Nat Conserv 20:364–373. https://doi.org/10.1016/j.jnc.2012.08.002
Isaac NJB, Pocock MJO (2015) Bias and information in biological records. Biol J Lin Soc 115:522–531
Isaac NJB et al (2020) Data integration for large-scale models of species distributions. Trends Ecol Evol 35:56–67
Johnston A, Hochachka WM, Strimas-Mackey ME, Gutierrez VR, Robinson OJ, Miller ET, Auer T, Kelling ST, Fink D (2019) Best practices for making reliable inferences from citizen science data: case study using eBird to estimate species distributions. bioRxiv:1–13
Johnston A, Moran N, Musgrove A, Fink D, Baillie SR (2020) Estimating species distributions from spatially biased citizen science data. Ecol Model 422:108927. https://doi.org/10.1016/j.ecolmodel.2019.108927
Juniper T, Parr M (1998) Parrots: a guide to parrots of the world. Helm, UK
Kai Z, Woan TS, Jie L, Goodale E, Kitajima K, Bagchi R, Harrison RD (2014) Shifting baselines on a tropical forest frontier: Extirpations drive declines in local ecological knowledge. PLoS ONE 9(1):86598
Kosmala M, Wiggins A, Swanson A, Simmons B (2016) Assessing data quality in citizen science. Front Ecol Environ 14:551–560
Lahoz-Monfort JJ, Guillera-Arroita G, Wintle BA (2014) Imperfect detection impacts the performance of species distribution models. Glob Ecol Biogeogr 23:504–515
Langham GM, Schuetz JG, Distler T, Soykan CU, Wilsey C (2015) Conservation status of north american birds in the face of future climate change:1–16
Lopes DC, Martin RO, Henriques M, Monteiro H, Cardoso PAULO, Tchantchalam Q, Pires AJ, Regalla A, Catry P (2018) Combining local knowledge and field surveys to determine status and threats to Timneh Parrots Psittacus timneh in Guinea-Bissau. Bird Conserv Int 2012:400–412
Mallory ML, Gilchrist HG, Fontaine AJ, Akearok JA (2003) Local ecological knowledge of ivory gull declines in Arctic Canada. Arctic 56:293–298
Melovski D et al (2018) Using questionnaire surveys and occupancy modelling to identify conservation priorities for the Critically Endangered Balkan lynx Lynx lynx balcanicus. Oryx 54(5):706–714
Mideros MA (2012) Ecuador: defining and measuring multidimensional poverty, 2006–2010. CEPAL Rev 2012:49–67
O’Brien TG, Winarni NL, Saanin FM, Kinnaird MF, Jepson P (1998) Distribution and conservation status of Bornean Peacock-pheasant Polyplectron schleiermacheri in Central Kalimantan, Indonesia. Bird Conserv Int 8:373–385
Pace DS et al (2019) An integrated approach for cetacean knowledge and conservation in the central Mediterranean Sea using research and social media data sources. Aquat Conserv Mar Freshwat Ecosyst 29:1302–1323
Peterson AT, Soberón J, Pearson RG, Anderson RP, Martínez-Meyer E, Nakamura M, Araújo MB (2011) Ecological Niches and Geographic Distributions (MPB-49)
Phillips SJ, Anderson RP, Schapire RE (2006) Maximum entropy modeling of species geographic distributions. Ecological Modelling 190:231–259. Available from https://linkinghub.elsevier.com/retrieve/pii/S030438000500267X
Phillips S, Dudík M (2008) Modeling of species distributions with Maxent: new extensions and a comprehensive evaluation. Ecography 31(2):161–175
Polfus JL, Heinemeyer K, Hebblewhite M (2014) Comparing traditional ecological knowledge and western science woodland caribou habitat models. J Wildl Manag 78:112–121
Ridgely RS, Greenfield PJ (2001a) The birds of ecuador: status, distribution and taxonomy. Helm, UK
Ridgely RS, Greenfield PJ (2001b) The birds of ecuador: field guide. Comstock Pub, USA
Robinson TP, Van Klinken RD, Metternicht G (2010) Comparison of alternative strategies for invasive species distribution 1 modeling 2 3. Ecol Model 221. Available from https://doi.org/10.1016/j.ecolmodel.2010.04.018. Accessed 10 Feb 2020
Saunders SP, Michel NL, Bateman BL, Wilsey CB, Dale K, LeBaron GS, Langham GM (2020) Community science validates climate suitability projections from ecological niche modeling. Ecol Appl 30:1–17
Skroblin A, Carboon T, Bidu G, Chapman N, Miller M, Taylor K, Taylor W, Game E, Wintle B (2021) Including indigenous knowledge in species distribution modeling for increased ecological insights. Conserv Biol 35(2):587–597
Steen VA, Elphick CS, Tingley MW (2019) An evaluation of stringent filtering to improve species distribution models from citizen science data. Divers Distrib 25:1857–1869
Sullivan BL et al (2014) The eBird enterprise: An integrated approach to development and application of citizen science. Biol Conserv 169:31–40. Elsevier Ltd. https://doi.org/10.1016/j.biocon.2013.11.003
Tjur T (2009) Coefficients of determination in logistic regression models - a new proposal: the coefficient of discrimination. Am Stat 63:366–372
Tourangeau R, Yan T (2007) Sensitive Questions in Surveys. Psychol Bull 133:859–883
Voeks RA (2007) Are women reservoirs of traditional plant knowledge? Gender, ethnobotany and globalization in northeast Brazil. Singap J Trop Geogr 28:7–20
Warren D, Glor R, Turelli M (2010) ENMTools: a toolbox for comparative studies of environmental niche models. Ecography 33:607–611
Warren DL, Seifert SN (2011) Ecological niche modeling in Maxent: the importance of model complexity and the performance of model selection criteria. Ecol Appl 21:335–342. Wiley. https://doi.org/10.1890/10-1171.1. Accessed 11 Mar 2020
Wickham H (2016) ggplot2: Elegant graphics for data analysis. Springer-Verlag, New York. Available from https://ggplot2.tidyverse.org
Wilson KA, Westphal MI, Possingham HP, Elith J (2005) Sensitivity of conservation planning to different approaches to using predicted species distribution data. Biol Cons 122:99–112
Wisz MS et al (2008) Effects of sample size on the performance of species distribution models. Divers Distrib 14:763–773
Zuur AF, Ieno EN, Elphick CS (2010) A protocol for data exploration to avoid common statistical problems. Methods Ecol Evol 1:3–14
Acknowledgements
Many thanks to all of the community members who volunteered their time to take part in our questionnaires; without them this research would not have been possible. Thank you to the North of England Zoological Society for providing all the funds to support this research.
Funding
This research was fully funded by the North of England Zoological Society.
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception. Material preparation and manuscript writing were conducted RB. Data collection was assisted by ISP, data analysis were performed by RB under the supervision of CD. MJ and SM commented on previous versions of the manuscript and provided conceptual guidance and critical revisions.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This study meets full compliance with ethical standards regarding research involving human participants. Full ethical approval for the questionnaires conducted was gained from the North of England Zoological Ethical Review Committee. Questionnaire content and an informed consent statement discussed with each participant prior to interviews, can be found in the online material.
Additional information
Communicated by Stephen Garnett.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Biddle, R., Solis-Ponce, I., Jones, M. et al. The value of local community knowledge in species distribution modelling for a threatened Neotropical parrot. Biodivers Conserv 30, 1803–1823 (2021). https://doi.org/10.1007/s10531-021-02169-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10531-021-02169-9