Abstract
Bacteria often release diverse iron-chelating compounds called siderophores to scavenge iron from the environment for many essential biological processes. In peatlands, where the biogeochemical cycle of iron and dissolved organic matter (DOM) are coupled, bacterial iron acquisition can be challenging even at high total iron concentrations. We found that the bacterium Pseudomonas sp. FEN, isolated from an Fe-rich peatland in the Northern Bavarian Fichtelgebirge (Germany), released an unprecedented siderophore for its genus. High-resolution mass spectrometry (HR-MS) using metal isotope-coded profiling (MICP), MS/MS experiments, and nuclear magnetic resonance spectroscopy (NMR) identified the amino polycarboxylic acid rhizobactin and a novel derivative at even higher amounts, which was named rhizobactin B. Interestingly, pyoverdine-like siderophores, typical for this genus, were not detected. With peat water extract (PWE), studies revealed that rhizobactin B could acquire Fe complexed by DOM, potentially through a TonB-dependent transporter, implying a higher Fe binding constant of rhizobactin B than DOM. The further uptake of Fe-rhizobactin B by Pseudomonas sp. FEN suggested its role as a siderophore. Rhizobactin B can complex several other metals, including Al, Cu, Mo, and Zn. The study demonstrates that the utilization of rhizobactin B can increase the Fe availability for Pseudomonas sp. FEN through ligand exchange with Fe-DOM, which has implications for the biogeochemical cycling of Fe in this peatland.
Similar content being viewed by others

Introduction
Siderophores are small weight molecules produced endogenously by microorganisms, fungi, or plants under iron (Fe)-limited conditions or taken up from the surrounding environment (Neilands 1981). Once siderophores are secreted into the environment, they contribute to the organism´s ligandosphere, which describes the entirety of excreted metal complexing agents and ligands derived from the DOM (Deicke et al. 2019). Currently, more than 500 siderophores produced by various plants, fungi, or microorganisms under Fe-limited conditions have been identified (Hider and Kong 2010), which can also be facilitated for the recruitment of various essential trace elements (Kraepiel et al. 2009). Siderophores can be categorized according to their moieties as catecholates, hydroxamates, carboxylates, and diazeniumdiolates (Ahmed and Holmstrom 2014; Hermenau et al. 2018). Early pioneering studies identified Fe-carriers with combined functional groups (Leong and Neilands 1982). For example, pyoverdine, produced by many members of the Pseudomonas genus, contains three parts, a dihydroxyquinoline core, an amino acid peptide chain that varies among strains, and a small dicarboxylic acid (or their monoamides). These substances often harbor a mix of hydroxamate and catecholate functional groups (Meyer 2000).
Pseudomonas spp. belong to a genus of gram-negative bacteria, which are ubiquitous in soils and aquatic ecosystems (Moore et al. 2006; Palleroni 1993). In these habitats, Fe and organic carbon are closely associated through the complexation with dissolved organic matter (DOM) or the formation of Fe-OM aggregates (Riedel et al. 2013). The Fe-DOM complexes and Fe-OM aggregates can thus control the accessibility of Fe for the microorganism, especially in carbon-rich habitats such as peatlands, despite the high Fe-concentrations (Kügler et al. 2019; Thomas Arrigo et al. 2016). Peatlands store approximately 30% of the land-based organic matter and thus more than any other vegetation type in the world (Bragazza et al. 2013; Mitra et al. 2005). DOM can stabilize both ferrous (FeII) and ferric (FeIII) Fe independent of the oxygen content in these environments (Bhattacharyya et al. 2018; Kügler et al. 2019). The co-occurrence of both Fe-redox states thus implies an essential role of microbial-mediated Fe-cycling in soils and peatlands. Both microbial FeII-oxidation and FeIII-reduction are enhanced in the presence of Fe-DOM complexes by either inhibiting abiotic FeII-oxidation or enhancing Fe-accessibility mediating electron transfer processes (Cooper et al. 2017; Hädrich et al. 2019; Kügler et al. 2019). Similarly, microbial Fe-assimilation mediated by siderophores might also be affected, as up to 97% of the Fe occurs in complexed form in peatlands (Kügler et al. 2019).
Our study thus aims to identify bacterial siderophores released by Pseudomonas spp. As a model organism, we used a Pseudomonas strain isolated from the Schlöppnerbrunnen fen, a well-studied peatland fed by FeII-rich groundwater in the Northern Bavarian Fichtelgebirge (Germany). Pseudomonas spp. are known to produce several other siderophores, including enantio-pyochelin (Youard et al. 2007), quinolobactin (Mossialos et al. 2000), ornicorrugatin (Matthijs et al. 2008) or pre-pseudomonine (Özkaya et al. 2015). Most of the Pseudomonas strains, such as P. fluorescens, produce one of these secondary siderophores in addition to the primary siderophore pyoverdine (Cornelis 2010). Pyoverdines are released under strong Fe-limitation, as they are very effective in Fe-recruitment (Dumas et al. 2013; Ross-Gillespie et al. 2015). Secondary siderophores can play interesting and unique roles in the biological or chemical activities of Pseudomonas spp., such as inflammation, biodegradation processes, as well as plant defense processes and antibiosis (Cornelis 2010; McRose et al. 2018). In this context, siderophores are also known to complex other metals such as Cu, Mo, W, or Zn for trace metal uptake (metallophores) or detoxification (Kraepiel et al. 2009; Johnstone and Nolan 2015; McRose et al. 2018; Wichard et al. 2008).
Over the past decades, several advances have been achieved to identify siderophores. Whereas improved spectrophotometric (Perez-Miranda et al. 2007) and microbial bioassays (Soria-Dengg et al. 2001) provide a tool for fast, mostly qualitative, siderophores-surveys, high-performance liquid chromatography/electrospray ionization mass spectrometry (HPLC–ESI–MS) based protocols were established for the quantification of known siderophores (Kilz et al. 1999; McCormack et al. 2003). The development of high-resolution mass spectrometry (HR-MS) using the natural Fe-isotopic pattern became a powerful tool to identify candidates of siderophores (Baars et al. 2016; Lehner et al. 2013). In this study, metal isotope-coded profiling (MICP) was applied to screen for unique isotopic signatures of the equally added amounts of Fe-isotopes, 54Fe, and 58Fe, in HR-MS (Deicke et al. 2014; Wichard 2016).
As the Fe-species of the fen soil are controlled by DOM (Kügler et al. 2019), the development of strategies for acquiring Fe from the peat water is essential for the peat microbial community. Thus, we hypothesized that our isolate, Pseudomonas sp. FEN, releases siderophores to manage the Fe-acquisition from the DOM. Using MICP, we screened the cell-free supernatant collected from Pseudomonas sp. FEN cultures grown under Fe-limited conditions for metallophores, and the structure of the isolated candidate siderophore was elucidated. Ligand exchange and short-term uptake experiments with Pseudomonas sp. FEN were performed to demonstrate the potential physiological function of the identified novel siderophore.
Materials and methods
Chemicals
Water, methanol, and acetonitrile for extraction and LC were UHPLC grade and purchased from VWR Chemicals (Darmstadt, Germany). MilliQ water (18.2 MΩ cm) was obtained from a MerckMillipore purification system (Darmstadt, Germany). Fe, Cu, Zn, and Mo-isotopes were purchased from Eurisotop (Saint-Aubin, France). Pyoverdine (from P. fluorescens strain ATCC 13,525) was obtained from EMC Microcollections (Tübingen, Germany). All other chemicals for growth media were purchased from Alfa Aesar (Ward Hill, MA, United States), AppliChem (Darmstadt, Germany), Roth (Karlsruhe, Germany) and Sigma Aldrich (Taufkirchen, Germany).
Sample material and cultivation
Pseudomonas sp. FEN was isolated from the Schlöppnerbrunnen fen in the Fichtelgebirge (Northern Bavaria, Germany) (Litzba 2009). Pseudomonas sp. FEN cultures were grown in PS medium overnight (ATCC medium 3: Nutrient broth medium) with shaking at room temperature. For incubation experiments, an aliquot (1 mL overnight 100 mL−1 media) of Pseudomonas sp. FEN overnight culture was transferred to serum bottles with modified Wolfe’s mineral medium (MWMM; ATCC medium 2672) containing 1 g NH4Cl, 0.2 g MgSO4·7H2O, 0.1 g CaCl2·2H2O, 0.05 g K2HPO4, amended with 10 mmol L−1 sodium lactate and 1 mL L−1 vitamin B solution (ATCC medium 2672). The pH of the media was adjusted to 6.5 with 10 mmol L−1 sodium hydrogen carbonate flushed with carbon dioxide before inoculation.
Growth curve of Pseudomonas sp. FEN
Triplicate Pseudomonas sp. FEN cultures were incubated in trace metal free MWMM (V = 100 mL) in serum bottles at 24 ± 1 °C for 42 h in a shaking incubator. To monitor the growth of Pseudomonas sp. FEN, optical density (OD600) was measured in polystyrene cuvettes (Sarstedt, Nümbrecht, Germany) using a UV–Vis spectrometer (Thermo Fischer Scientific, Waltham, USA). Each sample was measured in triplicate.
Phylogenetic analysis
Average nucleotide identity (ANI) values were calculated using the ANI-Matrix genome-based distance matrix calculator (Rodriguez-R and Konstantinidis 2016). The genome sequences for all Pseudomonas spp. were retrieved from the NCBI Assembly database in the form of nucleotide FASTA files, except for the newly sequenced Pseudomonas sp. FEN genome. Pseudomonas spp. strains used for the ANI comparisons were chosen based on the following two criteria: type of siderophore produced and the availability of the genome sequence in public databases. The ANI-matrix output was subsequently used for hierarchical clustering of the input genomes. The resulting tree is unrooted and derived using the BIONJ clustering method, a variant of the neighbor joining clustering method. The ANI-distance clustering tree, in Newick format, was visualized in MEGA X software (Stecher et al. 2020). Note, Pseudomonas cepacia ATCC25416 was renamed Burkholderia cepacia ATCC25416, and was used in the ANI-based analysis.
Genomic DNA extraction, genome sequencing and analysis
Pseudomonas sp. FEN cultures were grown in PS medium overnight with shaking at room temperature. Genomic DNA extraction, genome sequencing, and analysis were performed as described in Cooper et al. (2020). Briefly, biomass was harvested via centrifugation (10 min, 5000 × g, 4 °C) and genomic DNA was extracted using the GenElute Bacterial Genomic DNA kit (Sigma Aldrich, Taufkirchen) according to manufacturer’s instructions. Pseudomonas sp. FEN genomic DNA was used for whole-genome sequencing (PacBio sequencing), based on the standard manufacturer’s protocol (Cooper et al. 2020). The newly sequenced genome was annotated using RASTtk with default parameters (Aziz et al. 2008; Brettin et al. 2015; Overbeek et al. 2014). For analysis of the genome sequence, tonB- dependent receptor homologs were detected using BlastP (protein–protein BLAST) and the non-redundant protein sequences (nr) database.
Siderophore detection and identification using Chrome Azurol S and metal isotope-coded profiling
First, the chrome azurol S (CAS) assay, a universal colorimetric method, was applied in agar plates to detect siderophores independent of their structure. Siderophores scavenge Fe from the Fe-CAS-hexadecyltrimethylammonium bromide complex, and the free CAS dye changes from blue to yellow (Schwyn and Neilands 1987b). Agar plates prepared with PS medium were used to detect siderophore production by Pseudomonas sp. FEN and for the negative (abiotic) control.
To identify metallophores in the bacterial growth medium, cultures (V = 100 mL, OD600 = 0.2) were centrifuged (3500×g for 10 min at room temperature), and the supernatant was loaded on an HLB cartridge (225 mg, Oasis™, Waters, Milford, UK). Cartridges were conditioned with 3 mL methanol and equilibrated with 3 mL water, for solid-phase extraction (SPE). Compounds were eluted with 3 mL methanol (Deicke et al. 2014; Wichard 2016). The eluate was evaporated to dryness under nitrogen-steam and redissolved with 100 µL aqueous ammonium acetate (10 mmol L−1) at pH 6.6.
The identification of metal-binding complexes followed the workflow of metal isotope-coded profiling (MICP) identifying all peaks which show mass signals in a 1:1 intensity ratio and a mass difference of one of the applied isotope pairs (54Fe/58Fe = 3.9937u, 63Cu/65Cu = 1.9982u, 66Zn/68Zn = 1.9988u and 95Mo/98Mo = 2.9996u) (Deicke et al. 2014). Data was processed via DeltaMS, according to Baumeister et al. (2018). For example, to screen for siderophores, the Fe-isotopes 54Fe and 58Fe were solved in hydrochloric acid and were diluted with MilliQ water to 10–2 mol L−1. An isotope mixture containing equal amounts of 54Fe- and 58Fe-isotopes was prepared and added to the redissolved extract (10–4 mol L−1). The “isotope signature” type was chosen to identify isotopologues containing the isotopes 54Fe and 58Fe. The allowed variation of the isotope ratio was set to 10%. The following parameters were used for peak detection and DeltaMS processing: FWHM 30, steps 2, max 5, sntresh 5, step 0.01, mzdiffMatched 0.02, RTwindow 10, noiseCutoff 10, enriTol 0.1, varEQ false, numAtom 2, maxLT 4, ppmw 5, monoTol false, intChoice into, alpha 0.05, dpeak 12, compareOnlyDistros false.
Mass spectrometry and nuclear resonance spectroscopy
Mass spectrometry were performed using the qExactive Plus Orbitrap (Thermo Scientific, Bremen, Germany) coupled with an Ultimate 3000 (Dionex, Sunnyvale, CA, USA) UHPLC. A Zorbax SB-C8 HPLC-column (150 × 4.6 mm; 1.7 µm; Agilent Technologies, Santa Clara, CA, USA) was used. Acetonitrile spiked with 10% water and 1 mmol L−1 ammonium acetate (B) and water spiked with 2% acetonitrile and 1 mmol L−1 ammonium acetate (A) were used as eluents. The gradient was started at 100% A for 0.2 min, ramped to 50% A at 4 min. 100% B was achieved at 4.2 min and was held for one minute followed by a throwback to 100% A until 6 min. This was held for 30 s.
Electrospray ionization was conducted in both negative and positive mode (ESI-/ESI+). The spray voltage was set to 3,300 V in negative ionization mode and 3,000 V in positive ionization mode, respectively. Capillary temperature was amounted to 360 °C, sheath gas flow to 60, aux gas flow to 20, sweep gas flow to 5 and AGC-target to 3 × 106. Full scan analysis was conducted from m/z 100 to 1500 with a resolution of 70,000. The collision energy for MS/MS experiments, which were conducted in ESI + mode, was set to 30 eV at a resolution of 17,500 at m/z 200. The isolation window was set to 0.4 m/z for both compounds. All ion fragmentation of pyoverdine was executed in ESI+ mode with a resolution of 70,000. The collision energy was set to 75 eV, and the scan range from 50 to 750 m/z.
For nuclear magnetic resonance (NMR) analysis, purification of the siderophore candidates from 5 L culture medium using SPE was achieved by preparative HPLC as described in the next chapter, but without the addition of 58Fe. NMR-spectroscopy (1H,13C-HMBC and 1H,13C-HSQC) was performed with a 600 MHz Bruker Avance III using the residual resonance of the solvent D2O as an internal standard for reference.
Preparation of the 58Fe-labelled siderophores
For short-term uptake experiments of siderophore candidates and subsequent analysis by inductively coupled plasma mass spectrometry (ICP-MS), a 58Fe-complex was prepared. 58Fe (3 mmol L−1) was added to the solid phase extract of the Pseudomonas sp. FEN supernatant. This procedure prevented 56Fe from being bound by the siderophore candidate, which was essential for the uptake experiments. To remove the excess of 58Fe, the 58Fe-complex was purified using an Agilent 1100 series HPLC system (Agilent Technologies, Santa Clara, CA, USA) and a non-end-capped Nucleosil® C8 HPLC column (250 × 21 mm, 5 µm, Macherey–Nagel, Düren, Germany). Water spiked with 2% acetonitrile (A) and 100% acetonitrile (B) were used as eluents. The gradient was started at 100% A for 1 min and was changed to 80% until 30 min elapsed. The fraction from 15 to 16 min was collected, and the solvent was removed using the vacuum centrifuge RVC 2–25 CP plus (Christ, Osterode am Harz, Germany). UHPLC-HR-MS monitored purification success.
58Fe-uptake experiments
Triplicates of Pseudomonas sp. FEN cultures (V = 25 mL, OD600 = 0.2) were centrifuged, washed three times with MWMM, and the cell pellets were resuspended in 25 mL of fresh medium. 58FeCl3, 58Fe-EDTA, 58Fe-pyoverdine, and 58Fe-rhizobactin B were added to the medium (pH was adjusted to pH 6.5), incubated for 2 min, and passed through a 0.45 µm cellulose nitrate filter (Sartorius, Göttingen, Germany).
The bacteria on the filter were rinsed immediately with each 25 mL 0.1 mol L−1 EDTA / 0.05 mol L−1 oxalate solution and 25 mL water. Filters were digested with 2 mL nitric acid (70%) at 70 °C for 1 h. 500 µL of the tuning solution (Agilent Technologies, Santa Clara, CA, USA) containing 10 µg L−1 yttrium was added to 150 µL of the sample, then diluted to 5 mL with water and were measured via ICP-MS. The measured 58Fe-quota was corrected by the 58Fe-concentration measured on the filter treated with the corresponding Fe-species only (i.e., abiotic controls). This value was normalized to the background concentration of 58Fe in cultures not amended with an exogenous 58Fe source.
Inductively coupled plasma mass spectrometry
ICP-MS measurements were conducted to determine the concentration of 58Fe. The measurements were performed with the Agilent 7500c ICP-MS system (Agilent Technologies, Santa Clara, USA), equipped with a Babington nebulizer, a Scott spray chamber (cooled to 2 °C), and a Fassel torch. An ASX-500 autosampler (CETAC Technologies Inc., Ohama, USA) was connected to the ICP-MS with 1.02 cm ID PVC tubing for sample injection. The following settings were used for the ICP-MS measurements: RF power was set to 1,250 W, plasma gas flow set to 15 L min−1, and nebulizer gas flow set to 1.02 L min−1. The sample uptake time was 30 s (uptake speed: 350 µL min−1) and the rinse time 30 s.
Peat water extraction and metal determination
Peat cores were taken from the upper 30 cm of the Schlöppnerbrunnen fen, transported to the laboratory, and processed within 3 h for peat water extractions. Briefly, 130 g (wet weight) peat was added to 1 L autoclaved MilliQ water, and the slurry was shaken at 400 rpm for 24 h at 4 °C. After the 24 h incubation period, the peat water extract (PWE) was filtered through 0.3 μm glass fiber filters (Whatman, Maidstone, Great Britain) and either used immediately or stored at 4 °C until further use. For ligand exchange experiments with the purified siderophore, the ligand was added to the PWE and incubated for 10 min at 20 ± 1 °C and at the pH of the PWE (pH 5.5). The UHPLC-HR-MS measurements were immediately performed after the ligand exchange to compare the peak areas. The metal content of the PWE was determined using atomic absorption spectroscopy (AAS) for Fe and ICP-MS for Al, Cu, Mo and Zn, according to Kügler et al. (2019).
Monitoring of siderophore production during bacterial growth
For the growth-dependent production of rhizobactin B, 2 mL of the above-mentioned Pseudomonas sp. FEN cultures were sampled at the same time points as for the determination of the growth curve. SPE of the supernatant was performed using HLB cartridges (30 mg, Oasis™, Waters, Milford, UK), which were preconditioned with 1 mL methanol and equilibrated with 1 mL water before sample loading. Following the elution of rhizobactin B with 1 mL water, evaporating the water under nitrogen-stream, and dissolving the residue in water (40 µL), the purified rhizobactin B samples were analyzed by UHPLC-HR-MS.
Statistical analysis
Statistical tests, including paired t-test and Dunnet’s test, were performed with Microsoft Excel 2016 (Redmont, WA, USA) and Minitab 16 (Minitab Inc., State College, PA, USA), respectively.
Results
Siderophore production by Pseudomonas sp. FEN
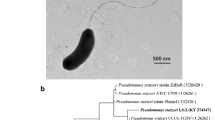
In order to evaluate the production and secretion of siderophores, Pseudomonas sp. FEN was grown in PS media to exponential phase and transferred to PS-CAS plates for direct detection and visualization of siderophores. The CAS assay revealed that Pseudomonas sp. FEN can produce siderophores, as indicated by a color change from blue to yellow and thus indicating a ligand exchange reaction between FeIII-CAS and competing siderophores (Fig. 1a).
Characterization of the siderophore producing members of the family Pseudomonadaceae. a Siderophore production of Pseudomonas sp. FEN grown on PS plates with CAS. Images of PS-CAS plates incubated for 24 h prior to photography indicate Pseudomonas sp. FEN produces siderophores, compared to the negative (abiotic) control, as indicated by the color change in PS-CAS plates inoculated with Pseudomonas sp. FEN. The CAS assay begins blue and changes to yellow as the ligand exchange reaction between FeIII-CAS and competing siderophores occurs. The experiment was performed in triplicate. b ANI-distance clustering tree, in Newick format, based on pairwise ANI comparisons between various siderophore-producing Pseudomonas strains, including Pseudomonas sp. FEN isolated from the Schlöppnerbrunnen fen. ANI values are indicated by circles placed on the respective node. Black circles designate ANI values > 90%, grey circles designate ANI values > 80%, and open circles designate ANI values < 80%. The scale bar refers to the ANI distance. Identified siderophores and the corresponding references are given if MS and/or NMR measurements were available
Taxonomic classification of Pseudomonas sp. FEN
To date, P. fluorescens strains include a larger group of pyoverdine producers, some non-producers, and those with various moieties binding metals. Initial 16S rRNA-based analysis indicated the Pseudomonas sp. FEN isolate was most closely related to P. fluorescens strains. However, recent advances in genomics and taxonomic classifications, namely the implementation of average nucleotide identity (ANI)-based comparisons and standardized taxonomy classifications (Genome Taxonomy Database taxonomy) (Lalucat et al. 2020), revealed our isolate is more closely related to the genome of type strain P. batumici, rather than any of the publicly available P. fluorescens genome sequences that comprise the P. fluorescens phylogenetic group within the P. fluorescens lineage of the Pseudomonadaceae genera (data not shown). This taxonomic classification of our isolate, Pseudomonas sp. FEN, further explains why this strain does not produce any fluorescent siderophores, such as pyoverdine, but instead can only utilize them. The Pseudomonas strains selected for comparison produce pyoverdines as well as secondary siderophores such as achromonobactin, azobactin, cepabactin, histicorrugatin, pyochelin, pseudomonine and rhizobactin-like siderophore and have publicly available genomes. Using these publicly available genomes along with the newly sequenced Pseudomonas sp. FEN genome, we were able to calculate the average nucleotide identities between all genomes selected. The ANI-based comparisons showed the Pseudomonas sp. FEN isolate is most similar to P. protegens Pf-5 and P. protegens CHA0. Additionally, the ANI-distance clustering tree revealed that assemblage of these diverse Pseudomonas strains appears to have branched evolutionarily to form two closely related subgroups (clusters) (Fig. 1b). Pseudomonas sp. FEN fits into an evolutionary cluster with P. savastanoi pv. phaseolicola 1448A, P. syringae pv. syringae B728a, P. syringae pv. tomato str. DC3000, B. cepacia ATCC 25,416, P. aeruginosa PAO1, P. entomophila L48, P. putida KT2440, P. putida GB-1, P. putida W15Oct28, P. protegens Pf-5 and P. protegens CHA0, while P. thivervalensis strain LMG 21,626, P. corrugata strain RM1-1–4, P. koreensis strain LMG 21,318, P. fluorescens Pf0-1, P. brenneri DSM 15,294, P. simiae strain PICF7, and P. fluorescens SBW25 fit into another cluster (Fig. 1b). Based on the ANI calculations, B. cepacia ATCC 25,416 has the lowest overall ANI percentages for all pairwise comparisons and the pairwise comparison between P. protegens Pf-5 and P. protegens CHA0 showed the highest ANI percentages (ANI 99%).
Siderophore screening
To identify Fe-chelating compounds, the solid phase extract of the supernatant of Pseudomonas sp. FEN grown under Fe-limited conditions was screened using HR-MS and DeltaMS (Fig. 2a, b). Fe-pyoverdine-complexes were detected neither in the exponential nor the stationary growth phase under Fe-limited conditions. Due to the structural variety of pyoverdines, we applied an all-ion-fragmentation to identify the characteristic fragments at m/z 204 and 230 for potential pyoverdines (Budzikiewicz et al. 2007). Using UHPLC-ESI–MS/MS, none of these fragments was detected in the extract of the supernatant of Pseudomonas sp. FEN culture. Moreover, no fluorescence, typical for pyoverdines, was measured in the filtered supernatant of the bacterial growth medium.
Identification of Fe-chelating compounds in Pseudomonas sp. FEN extract. a The total ion current (TIC, black line) and extracted ion chromatogram (EIC, blue and red lines) revealed two similar compounds, Fe-rhizobactin (1) (red line, intensity multiplied by factor 10) and elevated amounts of Fe-rhizobactin B (2) (blue line) detected by DeltaMS upon addition of stable pairs of the isotopes 54Fe and 58Fe in the positive electrospray ionization mode. Insert shows the readout of DeltaMS analysis at retention time 3.23 min: m/z 443.1186 and 447.1123. b The mass spectra reveals the characteristic isotopic signature of the quasi-molecule ions [M + H]+ of (1) m/z 429.1029; m/z 433.0965 and (2) m/z 443.1186; m/z 447.1123 (color code mentioned above). The additional peak pairs for (2) represent the CO2-depletion and the Na-adduct of rhizobactin B. c The corresponding EIC and mass spectra reveal the free ligands of both compounds under Fe-limited conditions
Due to the absence of pyoverdines, we used MICP to identify Fe-chelating compounds produced by the bacteria. The addition of the Fe-isotopes 54Fe and 58Fe revealed the presence of two sets of equally intense peaks in the mass spectrum with a mass difference of 3.9937 u, which was detected via DeltaMS in the Fe-limited Pseudomonas sp. FEN extract in the stationary phase (Fig. 2a). The HR-MS in ESI + mode indicated Fe-complexing compounds at m/z of 429.1028 and 433.0964 [M + H]+ as well as 443.1186 and 447.1123 [M + H]+ for the isotopes 54Fe and 58Fe, respectively. The other peak pairs represent the CO2-depletion and the Na-adduct of the pair at m/z 443 and 447 (Fig. 2a, b). The free ligands were identified as m/z 378.1866 [M + H]+ (1) and 392.2020 [M + H]+ (2) in the mass spectra without treatment of Fe-isotopes (Fig. 2c). The sum formula for the Fe-free ligand resulted in C15H27O8N3 (1) and C16H29O8N3 (2) based on the high-resolution mass, respectively. Both features depicted different but similar compounds due to the small retention time difference. HR-MS indicated that compound (2) possesses an additional methyl group in comparison to (1). Additional MS/MS experiments confirmed the structural relationship of both compounds showing a similar fragmentation pattern (Fig. 3a, b). The fragmentation pattern revealed the loss of two –COOH and two –C2H5N until m/z 200 for (1) and 214 for (2) for both compounds. Furthermore, there was the loss of one more carboxyl group and a visible C5H10N-backbone. The assigned sum formula for (1) and its fragmentation pattern lead to the assumption it depicted rhizobactin. Due to the similar fragmentation, the additional methyl-group for (2) could be localized in the suggested formula (Fig. 3c). Overall, Pseudomonas sp. FEN released two ligands complexing Fe, but not pyoverdine or any other siderophore typical for this genus. Siderophores such as enantio-pyochelin, quinolobactin, ornicorrugatin, or pre-pseudomonine were not identified by HR-MS under the applied conditions.
Mass spectrometry (MS/MS) analysis of the identified rhizobactin B (a) and rhizobactin (b). The structural formula indicates the fragmentation of rhizobactin. c The red moiety represents the varied fragment between both compounds. Illustration of fragment assignments of rhizobactin performed by an MS/MS experiment
Structure elucidation of rhizobactin B
To clarify the structural information of the rhizobactin derivative, 2D-NMR experiments were performed with the partly purified compound. In addition to the methyl group of alanine at δC 14.9 ppm, a second methyl-group was identified at δC 26.2 ppm (Table 1) which supports the mass spectrometric analysis. The chemical shift of δC 74.7 ppm and the corresponding 1H,13C-HSQC with no assigned proton indicated a quaternary carbon atom (C2) coupled to a hydroxyl group (Table 1). The quaternary carbon atoms C1 and C5 were assigned to a carboxyl and an amide group with chemical shifts of δC 181.6 and 173.1 ppm, respectively. Both carbon atoms C1 and C2 showed direct 1H,13C-HMBC couplings to H3, and H4, whereas C5 only depicted a coupling to H4. C/H3 represented a methyl group with the typical chemical shift of δH 1.30 ppm and δC 26.2 ppm. C/H4 displayed diastereomeric protons of a methylene-bridge at δH 2.44 and 2.64. Additionally, C3 showed an HMBC coupling with both H4 and C4 with H3. Therefore, we concluded that 1H,13C-HMBC, and 1H,13C-HSQC sufficiently revealed the position of the additional methyl-group at the quaternary C2 and belonged to the partial structure of citramalic acid (Fig. 4, Table 1).
Growth-dependent production of rhizobactin B and its function as a metallophore
The relative content of rhizobactin B in the supernatant peaked early in the lag phase (t-test, p < 0.05, n = 3) before it ceased when the exponential bacterial growth started (Fig. 5). Only traces of the Fe-complex were detected, due to the low Fe-concentration in the medium and the potential fast uptake by the bacterium. As production and release of rhizobactin B continued through the stationary phase, the highest rhizobactin B content was observed at the beginning of the stationary growth phase (t-test, p < 0.001, n = 3). The amount of rhizobactin B released during the remaining 17 h did not change significantly (t-test, p > 0.05, n = 3) (Fig. 5).
Growth of Pseudomonas sp. FEN under Fe-limited conditions. The growth of Pseudomonas sp. FEN over time was measured via optical density (OD600). The growth-dependent relative content of rhizobactin B (grey bar, in arbitrary units) was measured by UHPLC-HR-MS in the supernatant of the bacterial growth medium after solid-phase extraction at different time points. Error bars represent standard deviations of triplicate measurements
Further DeltaMS analysis revealed the complexation of other metals upon the addition of the isotopes 63Cu/65Cu, 66Zn/68Zn, and 95Mo/98Mo to the SPE of the Pseudomonas sp. FEN supernatant (Fig. 6a). In addition to Fe-complexation, mass spectra showed m/z pairs at 453.1161 and 455.1141 for Cu, 456.1135, and 458.1122 for Zn as well as 517.0823 and 520.0822 for Mo [M + H]+ (Fig. 6b). The assigned formulas were C16H27O8N3Cu, C16H27O8N3Zn, and C16H27O10N3Mo, which all belonged to the same ligand, rhizobactin B.
Metal complexation of rhizobactin B. a Extracted ion chromatograms (EIC) were created by plotting the intensity of the signal observed at the chosen m/z value of metal complexes. They show in vitro complexation of rhizobactin B after the addition of the stable pairs of isotopes 54Fe/58Fe, 63Cu/65Cu, 66Zn/68Zn and 95Mo/98Mo in a 1:1 ratio. b Corresponding mass spectra reveal the according mass shifts
Rhizobactin B acquires Fe from PWE and the Fe-complex is taken up by the bacterium
Furthermore, we tested whether rhizobactin B can acquire Fe and other metals from the PWE obtained from the Fe-rich Schlöppnerbrunnen fen. In addition to Fe, the PWE contained high amounts of Al and Zn, whereas Cu and Mo were negligible or even below the limit of detection (Table 2). After mixing the rhizobactin B with PWE, Al- and Zn-complexes could be identified in addition to Fe-rhizobactin B (Fig. 7a). As DOM entirely complexes Fe in PWE (Kügler et al. 2019), the change of the ratio between ligand and Fe-complex of rhizobactin B could be determined upon the addition of PWE. The stock solution of rhizobactin contained a 30-fold excess of the free ligand to the Fe-complex. After mixing PWE and rhizobactin B, the ratio changes in favor of the Fe-complex, and the amount of free ligand decreases to approximately 10% of the starting value (Fig. 7b).
Ligand exchange between rhiozobactin B and Fe-DOM. a Mass spectrometric analysis of the peat water extract (PWE) revealed metal complexes of rhizobactin B with Al, Fe, and Zn in the dissolved organic matter (DOM). b The decreasing ratio of free ligand to Fe-complex after the addition of PWE containing Fe-DOM confirms the ligand exchange of Fe (t-test, *** p < 0.001, n = 3). Error bars represent standard deviations of three measurements. c Uptake experiment of several 58Fe-sources (1.0 × 10–6 mol L−1) in Pseudomonas sp. FEN indicated increased Fe-uptake using rhizobactin B (Dunnett’s test in comparison with the control (white bar), p < 0.05, n = 3). Error bars represent standard deviations of triplicate measurements
Lastly, the physiological function of rhizobactin B as a siderophore for Fe-uptake was confirmed by short-term 58Fe uptake experiments with resuspended Pseudomonas sp. FEN cell pellets. Indeed, rhizobactin B doubled the Fe-uptake within the first 2 min of incubation (Dunnett’s test, *p < 0.05, n = 3) compared to the uptake of 58Fe-EDTA (control experiment), which was not significantly different from 1.0, which means no uptake (t-test, p > 0.05, n = 3) (Fig. 7c). Furthermore, we observed that the uptake of 58Fe-pyoverdine increased to 1.66 after 2 min, but it was not statistically significant due to the high variation of the uptake measurements.
Pseudomonas sp. FEN genome analysis
Using a draft genome assembly of Pseudomonas sp. FEN, we reviewed our chemical analysis and uptake experiments. Importantly, putative genes encoding for the biosynthesis of siderophores, Fe-transport systems, as well as Fe-siderophore sensor proteins and siderophore receptor proteins were found in the Pseudomonas sp. FEN genome. For example, genes are present in this genome encoding complete ferrichrome (a hydroxamate siderophore) uptake systems. Additionally, genes encoding various ‘siderophore uptake systems’ were annotated, indicating these systems play a role in the uptake of different siderophores from the surrounding environments, including pyoverdines. Interestingly, two genes encoding homologs to rhsAB/rhbAB were annotated as diaminobutyrate-2-oxoglutarate aminotransferase and diaminopimelate decarboxylase in the Pseudomonas sp. FEN genome. The rhsAB/rhbAB genes are components of the siderophore rhizobactin 1021 biosynthesis gene cluster (rhbABCDEF/rhsABCDEF) detected in Ensifer meliloti 2011 (formerly (Sino)Rhizobium meliloti) (Lynch et al. 2001). The presence of a gene homologous to those encoding the rhizobactin 1021 gene cluster indicates that this particular biosynthetic gene cluster is capable of producing unique siderophores structures that differ in both size and structure, including rhizobactin 1021 and rhizobactin B, in diverse microorganisms, such as E. meliloti 2011 and Pseudomonas sp. FEN.
Genes encoding ECF sigma factors and surface signaling systems, heme oxygenases, TonB-dependent hemin, ferrichrome receptors, and the piuCAB gene cluster, which encodes a Fe-uptake factor and a hydroxamate-type ferrisiderophore receptor, were also found in the Pseudomonas sp. FEN genome. Additionally, the genome encodes genes homologous to pitABC, a ferric Fe ABC transport system with Fe-binding, ATP-binding, and permease components, and EfeUOB, a ferrous Fe-transport system with a permease, a periplasmic protein along with a peroxidase component. Additionally, genes encoding TonB-dependent receptors in the Pseudomonas sp. FEN genome were identified by a manual search of the RAST annotated genome. Nine TonB-dependent receptor genes were identified, along with the closest homologs (Table 3). The homolog with the highest percent identity at the amino acid level was selected for comparison. In addition to TonB-dependent receptors, the Pseudomonas sp. FEN genome was examined for genes encoding ECF sigma factors located near the TonB-dependent receptors and genes encoding siderophore-related, iron-related, and heme-related protein families. In total, 4 ECF sigma factor-related genes and the closest homologs were identified (Table S1). The Pseudomonas sp. FEN genome contains genes for a variety of siderophore receptors, including ferrichrome receptors, ferrienterochelin receptors, colicin receptors, hemin receptors, and non-specific iron siderophore sensor and receptor proteins, as well as genes encoding both ferric and ferrous iron ABC-type transport systems (Tables 3, S1). Also, the genome contains genes encoding a pyoverdine ABC transporter system, suggesting this strain can utilize exogenous pyoverdine (RAST #2206; Table S1, Fig. 7b). Taken together, the draft genome of Pseudomonas sp. FEN provides additional corroborating evidence that this microorganism is capable of active siderophore production, import and export, as well as detection of exogenous siderophores.
Discussion
Rhizobactin was originally found in Ensifer meliloti DM4 (former Rhizobium meliloti), a nitrogen-fixing bacterium associated with legumes (Smith and Neilands 1984; Smith et al. 1985). It is structurally different to rhizobactin 1021, a hydroxamate siderophore produced under Fe-stress by Ensifer meliloti 2011 (Lynch et al. 2001). The biosynthetic parts of rhizobactin were described as an ethylenediamine group coupled with alanine, lysine, and malic acid (Fig. 3c) (Smith et al. 1985). We thus suggest that the malic acid in rhizobactin is replaced by citramalic acid for the biosynthesis of rhizobactin B, explaining the additional methyl group at C2 (Fig. 4, Table 1). Citramalic acid is known as a bacterial metabolite and building block (Khorassani et al. 2011). Indeed, P. fluorescens produces citramalic acid from itaconic acid (Cooper and Kornberg 1962; Nagai 1963).
In general, rhizobactin B belongs to a group of amino polycarboxylic acids, which are known to function as siderophores in bacteria or fungi (Drechsel et al. 1991; Meiwes et al. 1990). Until now, there was only an intermittently described connection between the production of amino polycarboxylic acids acting as siderophore and pseudomonads. Meyer and Hohnadel (1992) reported an increasing growth of several Pseudomonas strains in the presence of the xenobiotic nitrolotriacetic acid (NTA) due to the promotion of the Fe-uptake instead of utilizing NTA as carbon or nitrogen source. Interestingly, other similar amino polycarboxylic acids, such as ethylenediaminetetraacetic acid, inhibited Pseudomonas sp. growth (Meyer and Hohnadel 1992). Furthermore, genome analysis in recent studies of P. corrugata strain RM1-1-4 suggested genes involved in the production of rhizobactin-like siderophores (Zachow et al. 2017). The few overlaps between amino polycarboxylic acids and pseudomonads, as well as the missing pyoverdine production, illustrate the unique profile in the siderophore formation obtained for the Pseudomonas strain isolated from the fen. Furthermore, the presence of genes homologous to the rhizobactin 1021 biosynthetic gene cluster in the genome of Pseudomonas sp. FEN, as well as the clearly different structure of rhizobactin B compared to rhizobactin 1021, suggests that this biosynthetic gene cluster is used to produce unique siderophores by different microorganisms. However, the mechanisms regulating the production of the unique siderophore compounds produced by this gene cluster during rhizobactin biosynthesis is not known. Further studies will elucidate the biosynthesis of rhizobactin B by deeper analysis of the genome sequence and molecular biological approaches.
Interestingly, Pseudomonas sp. FEN began to release rhizobactin B in the early lag phase (Fig. 5), during which Fe was required for growth (Bellenger et al. 2011). In the exponential growth phase, the depletion of rhizobactin B in the growth medium indicates that the Fe-recruitment was faster than its secretion, which underlined the monitored Fe-uptake using rhizobactin B (Fig. 7c). Subsequently, we observed a substantial increase of rhizobactin B in the supernatant during the stationary phase. The reduced demand at the end of bacterial growth might explain the accumulation of the siderophore in the supernatant of the growth medium (Deicke et al. 2013).
Our results show that Pseudomonas sp. FEN can acquire Fe via the uptake of Fe-pyoverdine in addition to Fe-rhizobactin B (Fig. 7c). However, pyoverdines were not produced by Pseudomonas sp. FEN. The large structural variety of pyoverdines, especially in the amino acid chain structure or length, plays an essential role in the ability of microorganisms to utilize these primary siderophores, whether endogenously or exogenously produced. For instance, up to now, more than 50 pyoverdines of pseudomonads have been identified (Meyer et al. 2008). Strains of Pseudomonas were classified into (i) exclusive pyoverdine producers (Matthijs et al. 2009), (ii) pyoverdine producers that also form a secondary siderophore (Youard et al. 2007), (iii) exclusive secondary siderophore producers (Lewis et al. 2004) and (iv) strains without siderophore production (Champomier-Vergès et al. 1996). The inability to produce pyoverdines explains to why Pseudomonas sp. FEN does not cluster solely with the other P. fluorescens strains in the ANI-based clustering tree (Fig. 1b). Further studies are needed to explore the reasons for the lack of pyoverdine production in Pseudomonas sp. FEN, P. corrugata strain RM1-1-4 and P. syringae pv. tomato strain DC 3200. Up to now, only two different types of non-producers were described: those with a truncated pyoverdine locus and those with an intact, but silent locus (Butaitė et al. 2017). In addition, Hutchins et al. (1991) argued that the N-cost of siderophore production can be too high, therefore, the use of low molecular weight siderophores like rhizobactin B or even pyoverdines from an exogenous source might reflect a strategy to reduce metabolic costs for the biosynthesis of siderophores and Fe-uptake (Cornelis 2010; Matthijs et al. 2009). Siderophores are most useful as an Fe-recycling mechanism in more diffusion-controlled environments, where gradients of siderophore concentration enable its acquisition by a variety of coexisting microorganisms (Hutchins et al. 1991). In this context, Butaité et al. (2017) demonstrated that approximately 9% of the total pseudomonad communities in different soil and aquatic pond environments were not able to produce pyoverdines, however, these non-producers are capable of exploiting pyoverdines released by another organisms to acquire Fe (Butaite et al. 2017). Moreover, the non-pyoverdine producers occurred more frequently in the soil environments (Butaitė et al. 2018). As higher cell densities characterize soil environments compared to aquatic environments, the probability increases significantly of being surrounded by other siderophore producers (Ross-Gillespie et al. 2009; Scholz and Greenberg 2015). While the uptake of pyoverdine is reserved for pseudomonads only (Matthijs et al. 2009), they are flexible in their uptake of siderophores from other resources (Galet et al. 2015). Multiple Fe-uptake systems were identified in the genome of Pseudomonas sp. FEN (Table S1). Homologs to the TonB dependent receptor genes identified in the Pseudomonas sp. FEN genome which mediate the transport of siderophores were identified in P. asplenii, P. batumici or P. fluorescens (Table 3). TonB-dependent receptors mediate substrate-specific transport across the outer membrane, utilizing energy derived from the inner membrane complex TonB − ExbB − ExbD (Shultis et al. 2006), all of which were found in the Pseudomonas sp. FEN genome (Table 3). Several potential TonB systems might be involved in the uptake of siderophores by Pseudomonas sp. FEN and must be investigated further. Interestingly, we found some potential encoding TonB-dependent receptor sequences, which were too short to represent active receptors (data not shown). This reduction might be due to a selective pressure from the environment. A similar observation has been made in the case of P. aeruginosa isolated from cystic fibrosis lungs (Dingemans et al. 2014).
Previous studies suggest that pseudomonads capable of using various siderophores are more effective scavengers for Fe than those who rely solely on endogenous siderophores (Matthijs et al. 2009). Likewise, a study by Goldberg (2000) argued that the variability in the Fe-uptake and the ability to decrease the metabolic costs depict the high adaptability of pseudomonads, ultimately resulting in the colonization of diverse ecological niches. As Fe-DOM complexes provide the majority of soluble Fe in the Schlöppnerbrunnen fen (Kügler et al. 2019), a ligand exchange must occur to acquire Fe and to overcome Fe-limitations. For example, in the presence of tannic acids, A. vinelandii produced a higher amount of metallophores to gain V and Mo to sustain nitrogen fixation (Jouogo Noumsi et al. 2016). In addition to the presence of soluble Fe, PWE, which was filtered through 0.3 µm glass fiber filters, often contains colloidal Fe-forms. These colloids are also known to be associated with DOM (Neubauer et al. 2013; Stolpe et al. 2013). Overall, ligand-associated Fe-acquisition is necessary to overcome the obstacles faces by the microorganisms.
The formation constant of stable Fe-DOM reaches up to 1014 L mol−1 (Rue and Bruland 1995), whereas Fe-siderophores can form more stable complexes up to 1052 L mol−1, depending on the ligand (Kraemer 2004; Lalonde et al. 2012). Even though the smaller, secondary siderophores have a lower affinity for Fe (Matthijs et al. 2009), their formation constants are still larger than those of Fe-DOM. The relatively weak formation constant of the hard FeIII Lewis acid with rhizobactin is 1019 L mol−1 defined by the coordination to the comparatively soft 1,2-diaminoethane side and to the hard oxygen atoms of the carboxyl and hydroxyl groups (Schwyn and Neilands 1987a). These groups of rhizobactin can also coordinate to the borderline FeII acid (formation constant 109 L mol−1) according to the HSAB (hard and soft acids and bases) principle (Neilands 1993; Schwyn and Neilands 1987a). Here, we assume comparable binding properties for rhizobactin B. Indeed, rhizobactin B mixed with PWE outcompeted the DOM and chelated Fe for instant uptake (Fig. 7). Interestingly, although Pseudomonas mendocina could utilize Fe-DOM complexes even without the use of siderophores, they increase the availability of accessible Fe-species (Kuhn et al. 2012). We thus conclude that Fe-DOM complexes are readily available for the acquisition of Fe by siderophores as also previously reported (Kuhn and Maurice 2014).
As the metal complexation of rhizobactin B goes beyond Fe-complexation (Fig. 6), and elevated amounts of other metals were detected in the PWE of the Schlöppnerbrunnen fen (Table 2), it is tempting to assume that rhizobactin B also acquires, e.g., Zn from the metal-DOM pool under natural conditions or it is involved in detoxification processes. Nevertheless, the complexation of FeIII seems to be primary as the formation constants of typical metallophores, e.g., for ZnII, CuII and MnII-pyoverdine-complexes are much lower (1017 and 1022 L mol−1) compared to the constant of 1032 L mol−1 for the FeIII-pyoverdine-complex (Chen et al. 1994). Here, we assume similar changes in affinity for the metal complexation of rhizobactin B. The complexation of metals other than Fe, such as Cu, Mo, or Zn, is also in agreement with the mode of action of pyochelin, a secondary siderophore produced by P. aeruginosa (Braud et al. 2009; Cunrath et al. 2016), and recently identified metallophores in Frankia spp. (Deicke et al. 2019). In this context, we speculate that rhizobactin B and other metallophores can contribute to the metal homeostasis of the most dominating plants, e.g., the Molinia grasses, Sphagnum mosses and Carex sedges, found in the Schlöppnerbrunnen fen (Paul et al. 2006). For example, the phytosiderophore mugineic acid, which is released by graminaceous plants (Sugiura and Nomoto 1984) and structurally related to rhizobactin B, binds FeIII, NiII, CuII, or ZnII. This zincophore can be useful in recruiting Zn from the soil for Zn-deficient plants (Suzuki et al. 2006). Considering the ligandosphere, which describes the entirety of excreted metal complexing agents (Deicke et al. 2019), future studies will shed light on the interactions between sedges, bacteria and metal-DOM, which might be mediated by metal acquisition from the DOM in the fen.
Conclusion
The structural identification of siderophores still plays an essential role in the discovery of new natural products, which may be utilized for various medical or environmental applications. P. fluorescens strains are well-studied siderophore producers, which are known to produce fluorescent pyoverdines. However, the Pseudomonas sp. FEN, which was previously isolated from the Fe-rich Schlöppnerbrunnen fen, shows an unusual pattern in its siderophore production. This strain does not produce any pyoverdine under the studied conditions. Pseudomonas sp. FEN secreted a previously unknown (for this genus) Fe-chelating compound, the amino polycarboxylic acid rhizobactin B, utilized to overcome the Fe-limitations which microorganisms often face in nature. The Fe-uptake pattern of Pseudomonas sp. FEN using both rhizobactin B and pyoverdine indicates that this bacterium not only utilizes various mechanisms of Fe-acquisition, it also produces a unique variety of siderophores. These characteristics have the potential to enhance the Fe-accessibility and competitiveness of Pseudomonas sp. FEN against other microorganisms. The chelation of other metals (e.g., Zn) widens the ecological role of rhizobactin B for Pseudomonas sp. FEN, which will be investigated further in future studies.
Abbreviations
- ANI:
-
Average nucleotide identity
- AAS:
-
Atomic absorption spectroscopy
- CAS:
-
Chrome azurol S
- DOM:
-
Dissolved organic matter
- EDTA:
-
Ethylenediaminetetraacetic acid
- ESI:
-
Electrospray ionization
- HR-MS:
-
High-resolution mass spectrometry
- ICP-MS:
-
Inductively coupled plasma mass spectrometry
- MWMM:
-
Modified Wolfe’s mineral medium
- NMR:
-
Nuclear magnetic resonance spectroscopy
- MICP:
-
Metal isotope-coded profiling
- MS:
-
Mass spectrometry
- NTA:
-
Nitrolotriacetic acid
- PWE:
-
Peat water extract
- OD:
-
Optical density
- SPE:
-
Solid phase extraction
- UHPLC:
-
Ultra high-performance liquid chromatography
References
Ahmed E, Holmstrom SJM (2014) Siderophores in environmental research: roles and applications. Microb Biotechnol 7:196–208. https://doi.org/10.1111/1751-7915.12117
Aziz RK et al (2008) The RAST server: rapid annotations using subsystems technology. BMC Genomics. https://doi.org/10.1186/1471-2164-9-75
Baars O, Zhang X, Morel FMM, Seyedsayamdost MR (2016) The siderophore metabolome of Azotobacter vinelandii. Appl Environ Microbiol 82:27–39. https://doi.org/10.1128/AEM.03160-15
Baumeister TUH et al (2018) DeltaMS: a tool to track isotopologues in GC- and LC-MS data. Metabolomics 14:41. https://doi.org/10.1007/s11306-018-1336-x
Bellenger JP, Wichard T, Xu Y, Kraepiel AML (2011) Essential metals for nitrogen fixation in a free-living N2-fixing bacterium: chelation, homeostasis and high use efficiency. Environ Microbiol 13:1395–1411. https://doi.org/10.1111/j.1462-2920.2011.02440.x
Berti AD, Thomas MG (2009) Analysis of achromobactin biosynthesis by Pseudomonas syringae pv. syringae B728a. J Bacteriol 191:4594–4604. https://doi.org/10.1128/jb.00457-09
Bhattacharyya A, Schmidt MP, Stavitski E, Martínez CE (2018) Iron speciation in peats: chemical and spectroscopic evidence for the co-occurrence of ferric and ferrous iron in organic complexes and mineral precipitates. Org Geochem 115:124–137. https://doi.org/10.1016/j.orggeochem.2017.10.012
Bragazza L, Parisod J, Buttler A, Bardgett RD (2013) Biogeochemical plant-soil microbe feedback in response to climate warming in peatlands. Nat Clim Change 3:273–277. https://doi.org/10.1038/nclimate1781
Braud A, Hannauer M, Mislin GLA, Schalk IJ (2009) The Pseudomonas aeruginosa pyochelin-iron uptake pathway and its metal specificity. J Bacteriol 191:3517–3525. https://doi.org/10.1128/jb.00010-09
Brettin T et al (2015) RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci Rep. https://doi.org/10.1038/srep08365
Briskot G, Taraz K, Budzikiewicz H (1986) Pyoverdine type siderophores from Pseudomonas aeruginosa. Z Naturforsch C 41:497–506
Budzikiewicz H, Schäfer M, Fernandez DU, Matthijs S, Cornelis P (2007) Characterization of the chromophores of pyoverdins and related siderophores by electrospray tandem mass spectrometry. Biometals 20:135–144. https://doi.org/10.1007/s10534-006-9021-3
Butaite E, Baumgartner M, Wyder S, Kümmerli R (2017) Siderophore cheating and cheating resistance shape competition for iron in soil and freshwater Pseudomonas communities. Nat Commun 8:414. https://doi.org/10.1038/s41467-017-00509-4
Butaitė E, Kramer J, Wyder S, Kümmerli R (2018) Environmental determinants of pyoverdine production, exploitation and competition in natural Pseudomonas communities. Environ Microbiol 20:3629–3642. https://doi.org/10.1101/263004
Champomier-Vergès MC, Stintzi A, Meyer JM (1996) Acquisition of iron by the non-siderophore-producing Pseudomonas fragi. Microbiology 142:1191–1199. https://doi.org/10.1099/13500872-142-5-1191
Chen Y, Jurkevitch E, Barness E, Hadar Y (1994) Stability-constants of pseudobactin complexes with transition-metals. Soil Sci Soc Am J 58:390–396. https://doi.org/10.2136/sssaj1994.03615995005800020021x
Cooper RA, Kornberg HL (1962) Formation of pyruvate and acetyl-coenzyme A from itaconyl-coenzyme A by Pseudomonas sp. Biochim Biophys Acta 59:480–481. https://doi.org/10.1016/0006-3002(62)90202-0
Cooper RE, Eusterhues K, Wegner CE, Totsche KU, Küsel K (2017) Ferrihydrite-associated organic matter (OM) stimulates reduction by Shewanella oneidensis MR-1 and a complex microbial consortia. Biogeosciences 14:5171–5188. https://doi.org/10.5194/bg-14-5171-2017
Cooper RE, Wegner C-E, Küsel K (2020) Draft genome sequence of Pseudomonas sp. FEN, isolated from the Fe- and organic matter (OM)-rich Schlöppnerbrunnen fen. Microbiol Resour Announc (submitted)
Cornelis P (2010) Iron uptake and metabolism in pseudomonads. Appl Microbiol Biotechnol 86:1637–1645. https://doi.org/10.1007/s00253-010-2550-2
Cox CD, Graham R (1979) Isolation of an iron-binding compound from Pseudomonas aeruginoas. J Bacteriol 137:357–364. https://doi.org/10.1128/jb.137.1.357-364.1979
Cunrath O, Geoffroy VA, Schalk IJ (2016) Metallome of Pseudomonas aeruginosa: a role for siderophores. Environ Microbiol 18:3258–3267. https://doi.org/10.1111/1462-2920.12971
Deicke M, Bellenger J-P, Wichard T (2013) Direct quantification of bacterial molybdenum and iron metallophores with ultra-high-performance liquid chromatography coupled to time-of-flight mass spectrometry. J Chromatogr 1298:50–60. https://doi.org/10.1016/j.chroma.2013.05.008
Deicke M, Mohr JF, Bellenger J-P, Wichard T (2014) Metallophore mapping in complex matrices by metal isotope coded profiling of organic ligands. Analyst 139:6096–6099. https://doi.org/10.1039/c4an01461h
Deicke M, Mohr JF, Roy S, Herzsprung P, Bellenger J-P, Wichard T (2019) Metallophore profiling of nitrogen-fixing Frankia spp. to understand metal management in the rhizosphere of actinorhizal plants. Metallomics 11:810–821
Dingemans J et al (2014) The deletion of TonB-dependent receptor genes is part of the genome reduction process that occurs during adaptation of Pseudomonas aeruginosa to the cystic fibrosis lung. Pathog Dis 71:26–38. https://doi.org/10.1111/2049-632x.12170
Drechsel H, Metzger J, Freund S, Jung G, Boelaert JR, Winkelmann G (1991) Rhizoferrin: a novel siderophore from the fungus Rhizopus microporus var rhizopodiformis. Biol Met 4:238–243. https://doi.org/10.1007/bf01141187
Dumas Z, Ross-Gillespie A, Kümmerli R (2013) Switching between apparently redundant iron-uptake mechanisms benefits bacteria in changeable environments. Proc R Soc Lond Ser B 280:20131055. https://doi.org/10.1098/rspb.2013.1055
Galet J, Deveau A, Hôtel L, Frey-Klett P, Leblond P, Aigle B (2015) Pseudomonas fluorescens pirates both ferrioxamine and ferricoelichelin siderophores from Streptomyces ambofaciens. Appl Environ Microbiol 81:3132–3141. https://doi.org/10.1128/aem.03520-14
Goldberg JB (2000) Pseudomonas: global bacteria. Trends Microbiol 8:55–57. https://doi.org/10.1016/S0966-842X(99)01671-6
Hädrich A et al (2019) Microbial Fe(II) oxidation by Sideroxydans lithotrophicus ES-1 in the presence of Schlöppnerbrunnen fen-derived humic acids. FEMS Microbiol Ecol 94:fiz034. https://doi.org/10.1093/femsec/fiz034
Hermenau R et al (2018) Gramibactin is a bacterial siderophore with a diazeniumdiolate ligand system. Nat Chem Biol 14:841–843. https://doi.org/10.1038/s41589-018-0101-9
Hider RC, Kong X (2010) Chemistry and biology of siderophores. Nat Prod Rep 27:637–657. https://doi.org/10.1039/b906679a
Hutchins DA, Rueter JG, Fish W (1991) Siderophore production and nitrogenfixation are mutually exclusive strategies in Anabaena 712. Limnol Oceanogr 36:1–12. https://doi.org/10.4319/lo.1991.36.1.0001
Jouogo Noumsi C, Pourhassan N, Darnajoux R, Deicke M, Wichard T, Burrus V, Bellenger JP (2016) Effect of organic matter on nitrogenase metal cofactors homeostasis in Azotobacter vinelandii under diazotrophic conditions. Environ Microbiol Rep 8:76–84. https://doi.org/10.1111/1758-2229.12353
Johnstone TC, Nolan EM (2015) Beyond iron: non-classical biological functions of bacterial siderophores. Dalton Trans 44:6320–6339. https://doi.org/10.1039/c4dt03559c
Khorassani R, Hettwer U, Ratzinger A, Steingrobe B, Karlovsky P, Claassen N (2011) Citramalic acid and salicylic acid in sugar beet root exudates solubilize soil phosphorus. BMC Plant Biol 11:121. https://doi.org/10.1186/1471-2229-11-121
Kilz S, Lenz C, Fuchs R, Budzikiewicz H (1999) A fast screening method for the identification of siderophores from fluorescent Pseudomonas spp. by liquid chromatography electrospray mass spectrometry. J Mass Spectrom 34:281–290
Kraemer SM (2004) Iron oxide dissolution and solubility in the presence of siderophores. Aquat Sci 66:3–18. https://doi.org/10.1007/s00027-003-0690-5
Kraepiel AML, Bellenger JP, Wichard T, Morel FMM (2009) Multiple roles of siderophores in free-living nitrogen-fixing bacteria. Biometals 22:573–581. https://doi.org/10.1007/s10534-009-9222-7
Kügler S, Cooper RE, Wegner C-E, Mohr JF, Wichard T, Küsel K (2019) Iron-organic matter complexes accelerate microbial iron cycling in an iron-rich fen. Sci Total Environ 646:972–988
Kuhn KM, Dehner CA, Dubois JL, Maurice PA (2012) Iron acquisition from natural organic matter by an aerobic Pseudomonas mendocina bacterium: siderophores and cellular iron status. Geomicrobiol J 29:780–791. https://doi.org/10.1080/01490451.2011.619639
Kuhn KM, Maurice PA (2014) Accessibility of humic-associated Fe to a microbial siderophore: implications for bioavailability. Environ Sci Technol 48:1015–1022. https://doi.org/10.1021/es404186v
Lalonde K, Mucci A, Ouellet A, Gelinas Y (2012) Preservation of organic matter in sediments promoted by iron. Nature 483:198–200. https://doi.org/10.1038/nature10855
Lalucat J, Mulet M, Gomila M, Garcia-Valdes E (2020) Genomics in bacterial taxonomy: impact on the genus Pseudomonas. Genes 11:139. https://doi.org/10.3390/genes11020139
Lehner SM, Atanasova L, Neumann NKN, Krska R, Lemmens M, Druzhinina IS, Schuhmacher R (2013) Isotope-assisted screening for iron-containing metabolites reveals a high degree of diversity among known and unknown siderophores produced by Trichoderma spp. Appl Environ Microbiol 79:18–31. https://doi.org/10.1128/aem.02339-12
Leong SA, Neilands JB (1982) Siderophore production by phytopathogenic microbial species. Arch Biochem Biophys 218:351–359. https://doi.org/10.1016/0003-9861(82)90356-3
Lewis TA et al (2004) Physiological and molecular genetic evaluation of the dechlorination agent, pyridine-2,6-bis(monothiocarboxylic acid) (PDTC) as a secondary siderophore of Pseudomonas. Environ Microbiol 6:159–169. https://doi.org/10.1046/j.1462-2920.2003.00558.x
Litzba U (2009) Diversity and activity of iron-oxidizers and iron-reducers at oxic-anoxic interfaces. Bachelor thesis, Friedrich Schiller Universität Jena
Lynch D, O'Brien J, Welch T, Clarke P, Cuiv PO, Crosa JH, O'Connell M (2001) Genetic organization of the region encoding regulation, biosynthesis, and transport of rhizobactin 1021, a siderophore produced by Sinorhizobium meliloti. J Bacteriol 183:2576–2585. https://doi.org/10.1128/jb.183.8.2576-2585.2001
Matthijs S, Brandt N, Ongena M, Achouak W, Meyer JM, Budzikiewicz H (2016) Pyoverdine and histicorrugatin-mediated iron acquisition in Pseudomonas thivervalensis. Biometals 29:467–485. https://doi.org/10.1007/s10534-016-9929-1
Matthijs S, Budzikiewicz H, Schäfer M, Wathelet B, Cornelis P (2008) Ornicorrugatin, a new siderophore from Pseudomonas fluorescens AF76. Z Naturforsch C 63:8–12
Matthijs S, Laus G, Meyer JM, Abbaspour-Tehrani K, Schäfer M, Budzikiewicz H, Cornelis P (2009) Siderophore-mediated iron acquisition in the entomopathogenic bacterium Pseudomonas entomophila L48 and its close relative Pseudomonas putida KT2440. Biometals 22:951–964. https://doi.org/10.1007/s10534-009-9247-y
McCormack P, Worsfold PJ, Gledhill M (2003) Separation and detection of siderophores produced by marine bacterioplankton using high-performance liquid chromatography with electrospray ionization mass spectrometry. Anal Chem 75:2647–2652. https://doi.org/10.1021/ac0340105
McRose DL, Seyedsayamdost MR, Morel FMM (2018) Multiple siderophores: bug or feature? J Biol Inorg Chem 23:983–993. https://doi.org/10.1007/s00775-018-1617-x
Meiwes J, Fiedler HP, Haag H, Zahner H, Konetschnyrapp S, Jung G (1990) Isolation and characterization of Staphyloferrin A, a compound with siderophore activity from Staphylococcus hyicus DSM-20459. FEMS Microbiol Lett 67:201–205. https://doi.org/10.1111/j.1574-6968.1990.tb13863.x
Meyer J-M (2000) Pyoverdines: pigments, siderophores and potential taxonomic markers of fluorescent Pseudomonas species. Arch Microbiol 174:135–142. https://doi.org/10.1007/s002030000188
Meyer J-M, Gruffaz C, Raharinosy V, Bezverbnaya I, Schäfer M, Budzikiewicz H (2008) Siderotyping of fluorescent Pseudomonas: molecular mass determination by mass spectrometry as a powerful pyoverdine siderotyping method. Biometals 21:259–271. https://doi.org/10.1007/s10534-007-9115-6
Meyer J-M, Hohnadel D (1992) Use of nitrilotriacetic acid (NTA) by Pseudomonas species through iron metabolism. Appl Microbiol Biotechnol 37:114–118. https://doi.org/10.1007/bf00174214
Meyer J-M, Hohnadel D, Hallé F (1989) Cepabactin from Pseudomonas cepacia, a new type of siderophore. Microbiology 135:1479–1487. https://doi.org/10.1099/00221287-135-6-1479
Mitra S, Wassmann R, Vlek PLG (2005) An appraisal of global wetland area and its organic carbon stock. Curr Sci 88:25–35
Moon CD, Zhang XX, Matthijs S, Schäfer M, Budzikiewicz H, Rainey PB (2008) Genomic, genetic and structural analysis of pyoverdine-mediated iron acquisition in the plant growth-promoting bacterium Pseudomonas fluorescens SBW25. BMC Microbiol. https://doi.org/10.1186/1471-2180-8-7
Moore ERB, Tindall BJ, Martins Dos Santos VAP, Pieper DH, Ramos J-L, Palleroni NJ (2006) Nonmedical: Pseudomonas. In: Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E (eds) The Prokaryotes Proteobacteria: Gamma Subclass. Springer, New York, pp 646–703
Mossialos D et al (2000) Quinolobactin, a new siderophore of Pseudomonas fluorescens ATCC 17400, the production of which is repressed by the cognate pyoverdine. Appl Environ Microbiol 66:487–492. https://doi.org/10.1128/aem.66.2.487-492.2000
Nagai J (1963) Studies on itaconate metabolism 2. Citramalate metabolism in Pseudomonas fluorescens grown on itaconate. J Biochem 54:34–40. https://doi.org/10.1093/oxfordjournals.jbchem.a127743
Neilands JB (1981) Microbial iron compounds. Annu Rev Biochem 50:715–731. https://doi.org/10.1146/annurev.bi.50.070181.003435
Neilands JB (1993) Overview of bacterial iron transport and siderophore systems in rhizobia. In: Barton LL, Hemming BC (eds) Iron chelation in plants and soil microorganisms. Academic Press Inc, San Diego, pp 180–196
Neubauer E, Schenkeveld WDC, Plathe KL, Rentenberger C, von der Kammer F, Kraemer SM, Hofmann T (2013) The influence of pH on iron speciation in podzol extracts: iron complexes with natural organic matter, and iron mineral nanoparticles. Sci Total Environ 461–462:108–116. https://doi.org/10.1016/j.scitotenv.2013.04.076
Overbeek R et al (2014) The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res 42:D206–D214. https://doi.org/10.1093/nar/gkt1226
Owen JG, Ackerley DF (2011) Characterization of pyoverdine and achromobactin in Pseudomonas syringae pv. phaseolicola 1448a. BMC Microbiol 11:218. https://doi.org/10.1186/1471-2180-11-218
Özkaya FC, Bedir E, Hames EE (2015) A new siderophore from sponge associated Pseudomonas fluorescens 4.9.3. Rec Nat Prod 9:509–517
Palleroni NJ (1993) Pseudomonas classification: a new case-history in the taxonomy of gram-negative bacteria. Anton Leeuw Int J G 64:231–251
Parker DL et al (2014) Pyoverdine synthesis by the Mn(II)-oxidizing bacterium Pseudomonas putida GB-1. Front Microbiol. https://doi.org/10.3389/fmicb.2014.00202
Paul S, Küsel K, Alewell C (2006) Reduction processes in forest wetlands: tracking down heterogeneity of source/sink functions with a combination of methods. Soil Biol Biochem 38:1028–1039. https://doi.org/10.1016/j.soilbio.2005.09.001
Perez-Miranda S, Cabirol N, George-Tellez R, Zamudio-Rivera LS, Fernandez FJ (2007) O-CAS, a fast and universal method for siderophore detection. J Microbiol Methods 70:127–131. https://doi.org/10.1016/j.mimet.2007.03.023
Riedel T, Zak D, Biester H, Dittmar T (2013) Iron traps terrestrially derived dissolved organic matter at redox interfaces. Proc Natl Acad Sci USA 110:10101. https://doi.org/10.1073/pnas.1221487110
Rodriguez-R LM, Konstantinidis KT (2016) The enveomics collection: a toolbox for specialized analyses of microbial genomes and metagenomes. PeerJ 4:e1900v1. https://doi.org/10.7287/peerj.preprints.1900v1
Ross-Gillespie A, Dumas Z, Kümmerli R (2015) Evolutionary dynamics of interlinked public goods traits: an experimental study of siderophore production in Pseudomonas aeruginosa. J Evol Biol 28:29–39. https://doi.org/10.1111/jeb.12559
Ross-Gillespie A, Gardner A, Buckling A, West SA, Griffin AS (2009) Density dependence and cooperation: theory and a test with bacteria. Evolution 63:2315–2325. https://doi.org/10.1111/j.1558-5646.2009.00723.x
Rue EL, Bruland KW (1995) Complexation of iron(III) by natural organic ligands in the Central North Pacific as determined by a new competitive ligand equilibration adsorptive cathodic stripping voltammetric method. Mar Chem 50:117–138. https://doi.org/10.1016/0304-4203(95)00031-l
Scholz RL, Greenberg EP (2015) Sociality in Escherichia coli: enterochelin is a private good at low cell density and can be shared at high cell density. J Bacteriol 197:2122–2128. https://doi.org/10.1128/JB.02596-14
Shultis DD, Purdy MD, Banchs CN, Wiener MC (2006) Outer membrane active transport: structure of the BtuB:TonB complex. Science 312:1396–1399. https://doi.org/10.1126/science.1127694
Schwyn B, Neilands JB (1987a) Siderophores from agronomically important species of the Rhizobiacae. Comments Agric Food Chem 1:95–114
Schwyn B, Neilands JB (1987b) Universal chemical-assay for the detection and determination of siderophores. Anal Biochem 160:47–56. https://doi.org/10.1016/0003-2697(87)90612-9
Smith MJ, Neilands JB (1984) Rhizobaction, a siderophore from Rhizobium meliloti. J Plant Nutr 7:449–458. https://doi.org/10.1080/01904168409363211
Smith MJ, Shoolery JN, Schwyn B, Holden I, Neilands JB (1985) Rhizobactin, a structurally novel siderophore from Rhizobium meliloti. J Am Chem Soc 107:1739–1743. https://doi.org/10.1021/ja00292a047
Soria-Dengg S, Reissbrodt R, Horstmann U (2001) Siderophores in marine, coastal waters and their relevance for iron uptake by phytoplankton: experiments with the diatom Phaeodactylum tricornutum. Mar Ecol Prog Ser 220:73–82. https://doi.org/10.3354/meps220073
Stecher G, Tamura K, Kumar S (2020) Molecular evolutionary genetics analysis (MEGA) for macOS. Mol Biol Evol 37:1237–1239. https://doi.org/10.1093/molbev/msz312
Stolpe B, Guo L, Shiller AM (2013) Binding and transport of rare earth elements by organic and iron-rich nanocolloids in Alaskan rivers, as revealed by field-flow fractionation and ICP-MS. Geochim Cosmochim Acta 106:446–462. https://doi.org/10.1016/j.gca.2012.12.033
Sugiura Y, Nomoto K (1984) Phytosiderphores structures and properties of mugineic acid and their metal-complexes. Struct Bond 58:107–135. https://doi.org/10.1007/BFb0111313
Suzuki M et al (2006) Biosynthesis and secretion of mugineic acid family phytosiderophores in zinc-deficient barley. Plant J 48:85–97. https://doi.org/10.1111/j.1365-313X.2006.02853.x
Thomas Arrigo LK, Mikutta C, Lohmayer R, Planer-Friedrich B, Kretzschmar R (2016) Sulfidization of organic freshwater flocs from a minerotrophic peatland: speciation changes of iron, sulfur, and arsenic. Environ Sci Technol 50:3607–3616. https://doi.org/10.1021/acs.est.5b05791
Wichard T, Bellenger JP, Loison A, Kraepiel AML (2008) Catechol siderophores control tungsten uptake and toxicity in the nitrogen-fixing bacterium Azotobacter vinelandii. Environ Sci Technol 42:2408–2413. https://doi.org/10.1021/es702651f
Wichard T (2016) Identification of metallophores and organic ligands in the chemosphere of the marine macroalga Ulva (Chlorophyta) and at land-sea interfaces. Front Mar Sci 3:131. https://doi.org/10.3389/fmars.2016.00131
Wong-Lun-Sang S, Bernardini JJ, Hennard C, Kyslik P, Dell A, Abdallah MA (1996) Bacterial siderophores: structure elucidation, 2D 1H and 13C NMR assignments of pyoverdins produced by Pseudomonas fluorescens CHAO. Tetrahedron Lett 37:3329–3332. https://doi.org/10.1016/0040-4039(96)00569-2
Ye L et al (2014) Draft genome sequence analysis of a Pseudomonas putida W15Oct28 strain with antagonistic activity to gram-positive and Pseudomonas sp. pathogens. PLoS ONE 9:e110038. https://doi.org/10.1371/journal.pone.0110038
Youard ZA, Mislin GLA, Majcherczyk PA, Schalk IJ, Reimmann C (2007) Pseudomonas fluorescens CHA0 produces enantio-pyochelin, the optical antipode of the Pseudomonas aeruginosa siderophore pyochelin. J Biol Chem 282:35546–35553. https://doi.org/10.1074/jbc.M707039200
Zachow C, Müller H, Laireiter CM, Tilcher R, Berg G (2017) Complete genome sequence of Pseudomonas corrugata strain RM1-1-4, a stress protecting agent from the rhizosphere of an oilseed rape bait plant. Stand Genomic Sci 12:66. https://doi.org/10.1186/s40793-017-0278-7
Acknowledgements
This study was part of the Collaborative Research Centre Chemical Mediators in Complex Biosystems (CRC 1127 ChemBioSys) of the Friedrich Schiller University (FSU) Jena, funded by the Deutsche Forschungsgemeinschaft. We thank Jens D. Wurlitzer (Institute for Biodiversity, FSU Jena) for assistance in the laboratory, Carl-Eric Wegner (Institute for Biodiversity, FSU Jena) for helpful discussion and assistance with bioinformatics analysis, Peter Bellstedt (Institute for Organic Chemistry and Macromolecular Chemistry, FSU Jena) for providing NMR spectral data and Georg Pohnert for support and great discussions throughout this study (Institute for Inorganic and Analytical Chemistry, FSU Jena).
Funding
Open Access funding enabled and organized by Projekt DEAL. Open Access funding enabled and organized by Projekt DEAL. The Collaborative Research Centre (CRC 1127 ChemBioSys) of the Friedrich Schiller University Jena was funded by the Deutsche Forschungsgemeinschaft.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Kügler, S., Cooper, R.E., Boessneck, J. et al. Rhizobactin B is the preferred siderophore by a novel Pseudomonas isolate to obtain iron from dissolved organic matter in peatlands. Biometals 33, 415–433 (2020). https://doi.org/10.1007/s10534-020-00258-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10534-020-00258-w