Abstract
Purpose
Women diagnosed with breast cancer have heterogeneous survival outcomes that cannot be fully explained by known prognostic factors, and germline variation is a plausible but unconfirmed risk factor.
Methods
We used three approaches to test the hypothesis that germline variation drives some differences in survival: mortality loci identification, tumor aggressiveness loci identification, and whole-genome prediction. The 2954 study participants were women diagnosed with breast cancer before age 50, with a median follow-up of 15 years who were genotyped on an exome array. We first searched for loci in gene regions that were associated with all-cause mortality. We next searched for loci in gene regions associated with five histopathological characteristics related to tumor aggressiveness. Last, we also predicted 10-year all-cause mortality on a subset of 1903 participants (3,245,343 variants after imputation) using whole-genome prediction methods.
Results
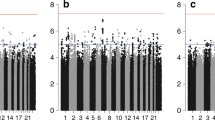
No risk loci for mortality or tumor aggressiveness were identified. This null result persisted when restricting to women with estrogen receptor-positive tumors, when examining suggestive loci in an independent study, and when restricting to previously published risk loci. Additionally, the whole-genome prediction model also found no evidence to support an association.
Conclusion
Despite multiple complementary approaches, our study found no evidence that mortality in women with early onset breast cancer is influenced by germline variation.


Similar content being viewed by others
Abbreviations
- AUC:
-
Area under the receiver operating characteristic
- CADD:
-
Combined annotation-dependent depletion
- EBI:
-
European Bioinformatics Institute
- ER:
-
Estrogen receptor
- GWAS:
-
Genome-wide association study
- HER2:
-
Human epidermal growth factor receptor 2
- NHGRI:
-
National Human Genome Research Institute
- PR:
-
Progesterone receptor
- QQ:
-
Quantile–quantile
- SKAT:
-
Sequence kernel association test
- SNV:
-
Single nucleotide variant
- TCGA:
-
The Cancer Genome Atlas
References
Nakano M, Fujisue M, Tashima R, Okumura Y, Nishiyama Y, Ohsako T, Toyozumi Y, Arima N, Nishimura R (2015) Survival time according to the year of recurrence and subtype in recurrent breast cancer. Breast 24(5):588–593
DeSantis CE, Fedewa SA, Goding Sauer A, Kramer JL, Smith RA, Jemal A (2016) Breast cancer statistics, 2015: convergence of incidence rates between black and white women. CA Cancer J Clin 66(1):31–42
Siegel RL, Miller KD, Jemal A (2017) Cancer statistics, 2017. CA Cancer J Clin 67(1):7–30
Tao L, Gomez SL, Keegan THM, Kurian AW, Clarke CA (2015) Breast cancer mortality in African- American and non-hispanic white women by molecular subtype and stage at diagnosis: a population based study. Cancer Epidemiol Biomark Prev 24(7):1039–1045
Menashe I, Anderson WF, Jatoi I, Rosenberg PS (2009) Underlying causes of the black-white racial disparity in breast cancer mortality: a population-based analysis. J Natl Cancer Inst 101(14):993–1000
Bai L, Yang HH, Hu Y, Shukla A, Ha NH, Doran A, Faraji F, Goldberger N, Lee MP, Keane T, Hunter KW (2016) An integrated genome-wide systems genetics screen for breast cancer metastasis susceptibility genes. PLoS Genet 12(4):e1005989
Lindström LS, Hall P, Hartman M, Wiklund F, Grönberg H, Czene K (2007) Familial concordance in cancer survival: a Swedish population-based study. Lancet Oncol 8(11):1001–1006
Hartman M, Lindström L, Dickman PW, Adami HO, Hall P, Czene K. Is breast cancer prognosis inherited? Breast Cancer Res 9(3):R39, 13
Lee S, Emond MJ, Bamshad MJ, Barnes KC, Rieder MJ, Nickerson DA, NHLBI GO Exome Sequencing Project ESP Lung Project Team, Christiani DC, Wurfel MM, Lin X (2012) Optimal unified approach for rare-variant association testing with application to small-sample case-control whole-exome sequencing studies. Am J Hum Genet 91(2):224–237
Schmidt M, Victor A, Bratzel D, Boehm D, Cotarelo C, Lebrecht A, Siggelkow W, Hengstler JG, Elsäÿer A, Gehrmann M, Lehr HA, Koelbl H, Minckwitz Gv, Harbeck N, Thomssen C (2009) Longterm outcome prediction by clinicopathological risk classification algorithms in node-negative breast cancercomparison between Adjuvant!, St Gallen, and a novel risk algorithm used in the prospective randomized Node-Negative-Breast Cancer-3 (NNBC-3) trial. Ann Oncol 20(2):258–264
Maas P, Barrdahl M, Joshi AD, et al (2016) Breast cancer risk from modifiable and nonmodifiable risk factors among white women in the United States. JAMA Oncol 2:1295–1302
Mavaddat N, Pharoah PDP, Michailidou K, Tyrer J, Brook MN, Bolla MK, Wang Q, Dennis J, Dunning AM, Shah M, Luben R, Brown J, Bojesen SE, Nordestgaard BG, Nielsen SF, Flyger H, Czene K, Darabi H, Eriksson M, Peto J, dos Santos-Silva I, Dudbridge F, Johnson N, Schmidt MK, Broeks A, Verhoef S, Rutgers EJ, Swerdlow A, Ashworth A, Orr N, Schoemaker MJ, Figueroa J, Chanock SJ, Brinton L, Lissowska J, Couch FJ, Olson JE, Vachon C, Pankratz VS, Lambrechts D, Wildiers H, Ongeval CV, Limbergen Ev, Kristensen V, Alnæs GG, Nord S, Borresen-Dale AL, Nevanlinna H, Muranen TA, Aittomäki K, Blomqvist C, Chang-Claude J, Rudolph A, Seibold P, Flesch-Janys D, Fasching PA, Haeberle L, Ekici AB, Beckmann MW, Burwinkel B, Marme F, Schneeweiss A, Sohn C, Trentham- Dietz A, Newcomb P, Titus L, Egan KM, Hunter DJ, Lindstrom S, Tamimi RM, Kraft P, Rahman N, Turnbull C, Renwick A, Seal S, Li J, Liu J, Humphreys K, Benitez J, Zamora MP, Perez JIA, Menéndez P, Jakubowska A, Lubinski J, Jaworska-Bieniek K, Durda K, Bogdanova NV, Antonenkova NN, Dörk T, Anton-Culver H, Neuhausen SL, Ziogas A, Bernstein L, Devilee P, Tollenaar RAEM, Seynaeve C, Asperen CJv, Cox A, Cross SS, Reed MWR, Khusnutdinova E, Bermisheva M, Prokofyeva D, Takhirova Z, Meindl A, Schmutzler RK, Sutter C, Yang R, Schürmann P, Bremer M, Christiansen H, Park-Simon TW, Hillemanns P, Guénel P, Truong T, Menegaux F, Sanchez M, Radice P, Peterlongo P, Manoukian S, Pensotti V, Hopper JL, Tsimiklis H, Apicella C, Southey MC, Brauch H, Brüning T, Ko YD, Sigurdson AJ, Doody MM, Hamann U, Torres D, Ulmer HU, Försti A, Sawyer EJ, Tomlinson I, Kerin MJ, Miller N, Andrulis IL, Knight JA, Glendon G, Mulligan AM, Chenevix-Trench G, Balleine R, Giles GG, Milne RL, McLean C, Lindblom A, Margolin S, Haiman CA, Henderson BE, Schumacher F, Marchand LL, Eilber U, Wang-Gohrke S, Hooning MJ, Hollestelle A, Ouweland AMWvd, Koppert LB, Carpenter J, Clarke C, Scott R, Mannermaa A, Kataja V, Kosma VM, Hartikainen JM, Brenner H, Arndt V, Stegmaier C, Dieenbach AK, Winqvist R, Pylkäs K, Jukkola-Vuorinen A, Grip M, O–t K, Vijai J, Robson M, Rau-Murthy R, Dwek M, Swann R, Perkins KA, Goldberg MS, Labrèche F, Dumont M, Eccles DM, Tapper WJ, Raq S, John EM, Whittemore AS, Slager S, Yannoukakos D, Toland AE, Yao S, Zheng W, Halverson SL, González-Neira A, Pita G, Alonso MR, Álvarez N, Herrero D, Tessier DC, Vincent D, Bacot F, Luccarini C, Baynes C, Ahmed S, Maranian M, Healey CS, Simard J, Hall P, Easton DF, Garcia-Closas M (2015) Prediction of breast cancer risk based on profiling with common genetic variants. J Natl Cancer Inst 107(5):djv036
Wheeler HE, Aquino-Michaels K, Gamazon ER, Trubetskoy VV, Dolan ME, Huang RS, Cox NJ, Im HK (2014) Poly-omic prediction of complex traits: OmicKriging. Genet Epidemiol 38(5):402415, arXiv: 1303.1788
The Cancer Genome Atlas Home Page. https://cancergenome.nih.gov/. Accessed 5 April 2017
Yang H, Wang K (2015) Genomic variant annotation and prioritization with ANNOVAR and wANNOVAR. Nat Protoc 10(10):15561566
Gibbs RA, Belmont JW, Hardenbol P, Willis TD, Yu F, Yang H, Ch’ang LY, Huang W, Liu B, Shen Y, Tam PKH, Tsui LC, Waye MMY, Wong JTF, Zeng C, Zhang Q, Chee MS, Galver LM, Kruglyak S, Murray SS, Oliphant AR, Montpetit A, Hudson TJ, Chagnon F, Ferretti V, Leboeuf M, Phillips MS, Verner A, Kwok PY, Duan S, Lind DL, Miller RD, Rice JP, Saccone NL, Taillon-Miller P, Xiao M, Nakamura Y, Sekine A, Sorimachi K, Tanaka T, Tanaka Y, Tsunoda T, Yoshino E, Bentley DR, Deloukas P, Hunt S, Powell D, Altshuler D, Gabriel SB, Zhang H, Matsuda I, Fukushima Y, Macer DR, Suda E, Rotimi CN, Adebamowo CA, Aniagwu T, Marshall PA, Matthew O, Nkwodimmah C, Royal CDM, Leppert MF, Dixon M, Stein LD, Cunningham F, Kanani A, Thorisson GA, Chakravarti A, Chen PE, Cutler DJ, Kashuk CS, Donnelly P, Marchini J, McVean GAT, Myers SR, Cardon LR, Abecasis GR, Morris A, Weir BS, Mullikin JC, Sherry ST, Feolo M, Daly MJ, Schaner SF, Qiu R, Kent A, Dunston GM, Kato K, Niikawa N, Knoppers BM, Foster MW, Clayton EW, Wang VO, Watkin J, Sodergren E, Weinstock GM, Wilson RK, Fulton LL, Rogers J, Birren BW, Han H, Wang H, Godbout M, Wallenburg JC, L’Archevêque P, Bellemare G, Todani K, Fujita T, Tanaka S, Holden AL, Lai EH, Collins FS, Brooks LD, McEwen JE, Guyer MS, Jordan E, Peterson JL, Spiegel J, Sung LM, Zacharia LF, Kennedy K, Dunn MG, Seabrook R, Shillito M, Skene B, Stewart JG, (chair) DLV, (co Chair) EWC, (co Chair) LBJ, Cho MK, Duster T, Jasperse M, Licinio J, Long JC, Ossorio PN, Spallone P, Terry SF, (chair) ESL, (co Chair) EHL, (co Chair) DAN, Boehnke M, Douglas JA, Hudson RR, Kruglyak L, Nussbaum RL (2003) The International HapMap Project. Nature 426(6968):789–796
The 1000 Genomes Project Consortium (2015) A global reference for human genetic variation. Nature 526(7571):68–74
Aulchenko YS, Ripke S, Isaacs A, van Duijn CM (2007) GenABEL: an R library for genome-wide association analysis. Bioinformatics 23(10):1294–1296
Karssen LC, van Duijn CM, Aulchenko YS (2016) The GenABEL Project for statistical genomics. F1000Research 5:914
R Core Team (2015) R: a language and environment for statistical computing, version3.2.2, 2015
Voorman A, Brody J, Lumley T (2013) skatMeta: efficient meta analysis for the SKAT test. https://cran.r-project.org/web/packages/skatMeta/index.html. Accessed 9 May 2017
Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, Shendure J (2014) A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet 46(3):310–315
Hu Q, Luo T, Zhong X, He P, Tian T, Zheng H (2015) Application status of tamoxifen in endocrine therapy for early breast cancer. Exp Therap Med 9(6):2207–2212
Muss HB (2001) Factors used to select adjuvant therapy of breast cancer in the United States: an overview of age, race, and socioeconomic status. JNCI Monogr 2001(30):52–55
Hertz DL, Rae JM (2016) One step at a time: CYP2D6 guided tamoxifen treatment awaits convincing evidence of clinical validity. Pharmacogenomics 17(8):823–826
Zembutsu H (2015) Pharmacogenomics toward personalized tamoxifen therapy for breast cancer. Pharmacogenomics 16(3):287–296
Kiyotani K, Mushiroda T, Tsunoda T, Morizono T, Hosono N, Kubo M, Tanigawara Y, Imamura CK, Flockhart DA, Aki F, Hirata K, Takatsuka Y, Okazaki M, Ohsumi S, Yamakawa T, Sasa M, Nakamura Y, Zembutsu H (2012) A genome-wide association study identifies locus at 10q22 associated with clinical outcomes of adjuvant tamoxifen therapy for breast cancer patients in Japanese. Hum Mol Genet 21(7):1665–1672
Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, Manolio TA (2009) Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci 106(23):93629367
Yang J, Lee SH, Goddard ME, Visscher PM (2011) GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet 88(1):7682
DeAbreu F, Schwartz G, Wells W, Tsongalis G (2014) Personalized therapy for breast cancer. Clin Genet 86(1):62–67
Bagaria SP, Ray PS, Sim MS, Ye X, Shamonki JM, Cui X, Giuliano AE (2014) Personalizing breast cancer staging by the inclusion of ER, PR, and HER2. JAMA Surg 149(2):125–129
Fletcher O, Johnson N, Orr N, Hosking FJ, Gibson LJ, Walker K, Zelenika D, Gut I, Heath S, Palles C, Coupland B, Broderick P, Schoemaker M, Jones M, Williamson J, Chilcott-Burns S, Tomczyk K, Simpson G, Jacobs KB, Chanock SJ, Hunter DJ, Tomlinson IP, Swerdlow A, Ashworth A, Ross G, dos Santos Silva I, Lathrop M, Houlston RS, Peto J (2011) Novel breast cancer susceptibility locus at 9q31.2: results of a genome-wide association study. J Natl Cancer Inst 103(5):425–435
Michailidou K, Hall P, Gonzalez-Neira A, Ghoussaini M, Dennis J, Milne RL, Schmidt MK, Chang- Claude J, Bojesen SE, Bolla MK, Wang Q, Dicks E, Lee A, Turnbull C, Rahman N (2013) The Breast and Ovarian Cancer Susceptibility Collaboration, Fletcher O, Peto J, Gibson L, dos Santos Silva I, Nevanlinna H, Muranen TA, Aittomäki K, Blomqvist C, Czene K, Irwanto A, Liu J, Waissz Q, Meijers- Heijboer H, Adank M, Hereditary Breast and Ovarian Cancer Research Group Netherlands (hebon), van der Luijt RB, Hein R, Dahmen N, Beckman L, Meindl A, Schmutzler RK, Müller-Myhsok B, Lichtner P, Hopper JL, Southey MC, Makalic E, Schmidt DF, Uitterlinden AG, Hofman A, Hunter DJ, Chanock SJ, Vincent D, Bacot F, Tessier DC, Canisius S, Wessels LFA, Haiman CA, Shah M, Luben R, Brown J, Luccarini C, Schoof N, Humphreys K, Li J, Nordestgaard BG, Nielsen SF, Flyger H, Couch FJ, Wang X, Vachon C, Stevens KN, Lambrechts D, Moisse M, Paridaens R, Christiaens MR, Rudolph A, Nickels S, Flesch-Janys D, Johnson N, Aitken Z, Aaltonen K, Heikkinen T, Broeks A, Veer LJV, van der Schoot CE, Guénel P, Truong T, Laurent-Puig P, Menegaux F, Marme F, Schneeweiss A, Sohn C, Burwinkel B, Zamora MP, Perez JIA, Pita G, Alonso MR, Cox A, Brock IW, Cross SS, Reed MWR, Sawyer EJ, Tomlinson I, Kerin MJ, Miller N, Henderson BE, Schumacher F, Le Marchand L, Andrulis IL, Knight JA, Glendon G, Mulligan AM, Investigators k, Group AOCS, Lindblom A, Margolin S, Hooning MJ, Hollestelle A, Ouweland AMWvd, Jager A, Bui QM, Stone J, Dite GS, Apicella C, Tsimiklis H, Giles GG, Severi G, Baglietto L, Fasching PA, Haeberle L, Ekici AB, Beckmann MW, Brenner H, Müller H, Arndt V, Stegmaier C, Swerdlow A, Ashworth A, Orr N, Jones M, Figueroa J, Lissowska J, Brinton L, Goldberg MS, Labrèche F, Dumont M, Winqvist R, Pylkäs K, Jukkola-Vuorinen A, Grip M, Brauch H, Hamann U, Brüning T, Network TGGEIaBCiG, Radice P, Peterlongo P, Manoukian S, Bonanni B, Devilee P, Tollenaar RAEM, Seynaeve C, Asperen CJv, Jakubowska A, Lubinski J, Jaworska K, Durda K, Mannermaa A, Kataja V, Kosma VM, Hartikainen JM, Bogdanova NV, Antonenkova NN, Dörk T, Kristensen VN, Anton-Culver H, Slager S, Toland AE, Edge S, Fostira F, Kang D, Yoo KY, Noh DY, Matsuo K, Ito H, Iwata H, Sueta A, Wu AH, Tseng CC, Berg DVD, Stram DO, Shu XO, Lu W, Gao YT, Cai H, Teo SH, Yip CH, Phuah SY, Cornes BK, Hartman M, Miao H, Lim WY, Sng JH, Muir K, Lophatananon A, Stewart-Brown S, Siriwanarangsan P, Shen CY, Hsiung CN, Wu PE, Ding SL, Sangrajrang S, Gaborieau V, Brennan P, McKay J, Blot WJ, Signorello LB, Cai Q, Zheng W, Deming-Halverson S, Shrubsole M, Long J, Simard J, Garcia-Closas M, Pharoah PDP, Chenevix-Trench G, Dunning AM, Benitez J, Easton DF. Large-scale genotyping identifies 41 new loci associated with breast cancer risk. Nat Genet 45(4):353–361
Ahsan H, Halpern J, Kibriya MG, Pierce BL, Tong L, Gamazon E, McGuire V, Felberg A, Shi J, Jasmine F, Roy S, Brutus R, Argos M, Melkonian S, Chang-Claude J, Andrulis I, Hopper JL, John EM, Malone K, Ursin G, Gammon MD, Thomas DC, Seminara D, Casey G, Knight JA, Southey MC, Giles GG, Santella RM, Lee E, Conti D, Duggan D, Gallinger S, Haile R, Jenkins M, Lindor NM, Newcomb P, Michailidou K, Apicella C, Park DJ, Peto J, Fletcher O, Silva IdS, Lathrop M, Hunter DJ, Chanock SJ, Meindl A, Schmutzler RK, Müller-Myhsok B, Lochmann M, Beckmann L, Hein R, Makalic E, Schmidt DF, Bui QM, Stone J, Flesch-Janys D, Dahmen N, Nevanlinna H, Aittomäki K, Blomqvist C, Hall P, Czene K, Irwanto A, Liu J, Rahman N, Turnbull C, Study ftFBC, Dunning AM, Pharoah P, Waissz Q, Meijers-Heijboer H, Uitterlinden AG, Rivadeneira F, Nicolae D, Easton DF, Cox NJ, and Whittemore AS. A genome-wide association study of early-onset breast cancer identifies PFKM as a novel breast cancer gene and supports a common genetic spectrum for breast cancer at any age. Cancer Epidemiol Biomark Prev 23(4):658–669
Fehringer G, Kraft P, Pharoah PD, Eeles RA, Chatterjee N, Schumacher FR, Schildkraut JM, Lindström S, Brennan P, Bickeböller H, Houlston RS, Landi MT, Caporaso N, Risch A, Amin Al Olama A, Berndt SI, Giovannucci EL, Grönberg H, Kote-Jarai Z, Ma J, Muir K, Stampfer MJ, Stevens VL, Wiklund F, Willett WC, Goode EL, Permuth JB, Risch HA, Reid BM, Bezieau S, Brenner H, Chan AT, Chang- Claude J, Hudson TJ, Kocarnik JK, Newcomb PA, Schoen RE, Slattery ML, White E, Adank MA, Ahsan H, Aittomäki K, Baglietto L, Blomquist C, Canzian F, Czene K, Dos-Santos-Silva I, Eliassen AH, Figueroa JD, Flesch-Janys D, Fletcher O, Garcia-Closas M, Gaudet MM, Johnson N, Hall P, Hazra A, Hein R, Hofman A, Hopper JL, Irwanto A, Johansson M, Kaaks R, Kibriya MG, Lichtner P, Liu J, Lund E, Makalic E, Meindl A, Müller-Myhsok B, Muranen TA, Nevanlinna H, Peeters PH, Peto J, Prentice RL, Rahman N, Sanchez MJ, Schmidt DF, Schmutzler RK, Southey MC, Tamimi R, Travis RC, Turnbull C, Uitterlinden AG, Wang Z, Whittemore AS, Yang XR, Zheng W, Buchanan DD, Casey G, Conti DV, Edlund CK, Gallinger S, Haile RW, Jenkins M, Le Marchand L, Li L, Lindor NM, Schmit SL, Thibodeau SN, Woods MO, Rafnar T, Gudmundsson J, Stacey SN, Stefansson K, Sulem P, Chen YA, Tyrer JP, Christiani DC, Wei Y, Shen H, Hu Z, Shu XO, Shiraishi K, Takahashi A, Bossé Y, Obeidat M, Nickle D, Timens W, Freedman ML, Li Q, Seminara D, Chanock SJ, Gong J, Peters U, Gruber SB, Amos CI, Sellers TA, Easton DF, Hunter DJ, Haiman CA, Henderson BE, Hung RJ, Ovarian Cancer Association Consortium (OCAC), PRACTICAL Consortium, Hereditary Breast and Ovarian Cancer Research Group Netherlands (HEBON), Colorectal Transdisciplinary (CORECT) Study, and African American Breast Cancer Consortium (AABC) and African Ancestry Prostate Cancer Consortium (AAPC). Cross-cancer genome-wide analysis of lung, ovary, breast, prostate, and colorectal cancer reveals novel pleiotropic associations. Cancer Res 76(17):5103–5114
Bahcall O (2013) Common variation and heritability estimates for breast, ovarian and prostate cancers. Nat Genet. http://www.nature.com/icogs/primer/common-variation-and-heritability-estimates-for-breast-ovarian-and-prostate-cancers/. Accessed 9 May 2017
Evans DG, Astley S, Stavrinos P, Harkness E, Donnelly LS, Dawe S, Jacob I, Harvie M, Cuzick J, Brentnall A, Wilson M, Harrison F, Payne K, Howell A (2016) Improvement in risk prediction, early detection and prevention of breast cancer in the NHS Breast Screening Programme and family history clinics: a dual cohort study. Programme Grants for Applied Research. NIHR Journals Library, Southampton (UK)
Fagerholm R, Khan S, Schmidt MK, García-Closas M, Heikkilä P, Saarela J, Beesley J, Jamshidi M, Aittomäki K, Liu J, Ali HR, Andrulis IL, Beckmann MW, Behrens S, Blows FM, Brenner H, Chang-Claude J, Couch FJ, Czene K, Fasching PA, Figueroa J, Floris G, Glendon G, Guo Q, Hall P, Hallberg E, Hamann U, Holleczek B, Hooning MJ, Hopper JL, Jager A, Kabisch M, kConFab/AOCS Investigators, Keeman R, Kosma VM, Lambrechts D, Lindblom A, Mannermaa A, Margolin S, Provenzano E, Shah M, Southey MC, Dennis J, Lush M, Michailidou K, Wang Q, Bolla MK, Dunning AM, Easton DF, Pharoah PDP, Chenevix-Trench G, Blomqvist C, Nevanlinna H (2017) TP53-based interaction analysis identifies cis-eQTL variants for TP53BP2, FBXO28, and FAM53A that associate with survival and treatment outcome in breast cancer. Oncotarget 8:18381–18398
Bardou VJ, Arpino G, Elledge RM, Osborne CK, Clark GM (2003) Progesterone receptor status significantly improves outcome prediction over estrogen receptor status alone for adjuvant endocrine therapy in two large breast cancer databases. J Clin Oncol 21(10):1973–1979
Acknowledgements
Portions of this manuscript have been included in the doctoral thesis of Molly Scannell Bryan at the University of Chicago (under embargo until 2019). The authors would like to thank Regina M. Santella of Columbia University. The content of this manuscript does not necessarily reflect the views or policies of the National Cancer Institute or any of the collaborating centers in the Breast Cancer Family Registry (BCFR), nor does mention of trade names, commercial products, or organizations imply endorsement by the USA Government or the BCFR. Samples from the CPIC were processed and distributed by the Coriell Institute for Medical Research. The contents of this manuscript are solely the responsibility of the authors and do not necessarily represent the official views of the National Cancer Institute.
Funding
This study was supported by the National Institute of Health Grants R01CA122171, RC1CA145506, U01CA122171, R01CA094069, UM1CA164920, RFA-CA-95-011, UO1CA/ES66572, UO1CA66572, U19CA148065, and R25-CA057699.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest. All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional research committee, and with the 1964 Helsinki declaration and its later amendments. Informed consent was obtained from all individual participants included in the study.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Scannell Bryan, M., Argos, M., Andrulis, I.L. et al. Limited influence of germline genetic variation on all-cause mortality in women with early onset breast cancer: evidence from gene-based tests, single-marker regression, and whole-genome prediction. Breast Cancer Res Treat 164, 707–717 (2017). https://doi.org/10.1007/s10549-017-4287-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-017-4287-4