Abstract
The citrus bark cracking viroid (CBCVd) was identified as causal agent for a severe stunting disease in hops. Viroids are highly stable parasitic RNAs, which can be easily transmitted by agricultural practices. Since CBCVd has recently been detected in two European countries a growing concern is that this pathogen will further spread and thereby threaten the European hop production. Biogas fermentation is used to sanitize hop harvest residues infected with pathogenic fungi. Consequently, the aim of this study was to test if biogas fermentation can contribute to viroid degradation at mesophilic (40 °C) and thermophilic (50 °C) conditions. Therefore, a duplex reverse transcription real-time PCR analysis was developed for CBCVd and HLVd detection in biogas fermentation residues. The non-pathogenic hop latent viroid (HLVd) was used as viroid model for the pathogenic CBCVd. The fermentation trials showed that HLVd was significantly degraded after 30 days at mesophilic or after 5 days at thermophilic conditions, respectively. However, sequencing revealed that HLVd was not fully degraded even after 90 days. The incubation of hop harvest residues at different temperatures between 20 and 70 °C showed that 70 °C led to a significant HLVd degradation after 1 day. In conclusion, we suggest combining 70 °C pretreatment and thermophilic fermentation for efficient viroid decontamination.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Viroids are small parasitic RNAs that replicate in their host plants by interacting with the host transcriptional machinery (Škorić, 2017). The viroid infections encompass the entire spectrum of disease intensity from only moderate as for example the infection of the hop latent viroid in hops (Jakse et al., 2015) to severe symptom expression caused by potato spindle tuber viroid (PSTVd) in potatoes (Diener, 1972). In certain cases, moderate symptoms like dwarfing might even be considered as beneficial. For example, so called “dwarfing viroids” are used for high-density plantings of citrus (Vidalakis et al., 2011). The citrus bark cracking viroid (CBCVd, synonym Citrus viroid IV) is one of those dwarfing viroids (Bar-Joseph, 1993). However, the CBCVd is also the causal agent for a severe hop stunting disease, which was first discovered in Slovenia in 2007 (Jakse et al., 2015). Until today this severe hop stunting disease has caused high economic losses and is still spreading (Radisek, 2017). The eradication of viroids from an agricultural production site is difficult because any plant material harboring viroids left on the field might act as source for further infection. Viroids easily spread by any practices that cause mechanical damage on plants, especially pruning and harvest (Škorić, 2017). Further, viroids can persist a certain time in water, in plant residues or even in compost (Kerins et al., 2018; Mehle et al., 2014). However, for viroids generally soil-born infection may not be a major risk (Seigner et al., 2008; Verhoeven et al., 2010). Nevertheless viroid transmission via roots is possible under experimental conditions and may also be realistic under practical conditions when plants are cultivated over long periods (Matsushita, 2013; Mehle et al., 2014). Consequently, hop growers have the concern that hop harvest residues used as fertilizers might still contain infectious viroid particles despite ensiling and fermentation, thus may cause infections. The primary infection of hop plants in Slovenia is thought to be the result of establishing a hop garden on an illegal refuse dump containing citrus plant residues (Radisek, 2017). Therefore, it seems possible that viroid-containing plant residues lead to (re-)infections. Distribution of hop harvest residues in hop gardens is a common practice not just for increasing soil fertility, but also as a solution for the disposal of the huge amount of above-ground plant material resulting from the annual hop harvest. Thus, sanitization by fermentation of hop harvest residues has been established in some hop production areas. In addition, farms with biogas plants are always looking for by-products that are suitable as substrate for their biogas plant to produce renewable energy. At the same time, hygienisation can take place via this utilization route (Oechsner, 2017). The new threat of CBCVd could in turn compromise this recycling strategy. A current study on the fate of the HLVd during ensiling could not resolve those concerns, because ensiling did not reduce HLVd quantities even after a period of 4 month (Hagemann et al., 2021). Therefore, the first aim of this study was to analyze if the fermentation processes in a biogas plant can lead to viroid degradation and subsequently can be used for sanitization of infected material. Generally, viroids are highly tolerant to thermal inactivation under experimental conditions as shown for the hop stunt viroid (HSVd), which was still able to infect plants after heat treatment for 10 min at 140 °C (Takahashi & Yaguchi, 1985). In contrast, in more practical oriented experiments with composting of PSTVd infected plant material at a temperature of 50 °C it was shown that viroid levels drop below detection limits within 2 weeks (Kerins et al., 2018). Thus, the matrix-temperature interaction and concomitant microbiological processes seem to be the crucial factor for viroid degradation. As consequence the aim was to study viroid decay in the biogas plant. Therefore, two typical temperature regimes were compared, mesophilic fermentation at 40 °C and thermophilic fermentation at 50 °C. Additionally, to study the temperature dependency of viroid degradation HLVd infected hop material was incubated in vitro at different temperatures and the secondary structure was predicted in silico accordingly.
The CBCVd has been evaluated by an express risk assessment by the European Plant Protection Organization (EPPO) (Radisek, 2017) and is likely to be listed as regulated non-quarantine pests (RNQP). To avoid any risk of dispersal of the dangerous CBCVd during the experiments, the closely related hop latent viroid (HLVd) has been used as model. CBCVd and HLVd both can infect hop plants and belong to the family of Cocadviroidae as shown by phylogenetic analysis (Di Serio et al., 2014). HLVd has been detected in all hop production areas worldwide and no serious HLVd-related pathogenesis is known for crops produced in the hop production areas. However, RNA sequencing of cannabis plants (Cannabis sativa L.) with and without stunting symptoms suggested that the HLVd is the agent causing a severe stunting disease in cannabis (Bektaş et al., 2019; Warren et al., 2019). Thus, this research might have direct implications for a HLVd inactivationmethods as part of cannabis harvest decontamination strategy. Analysis of the HLVd and the CBCVd are typically performed starting with a reverse transcription of the viroid RNA followed by a duplex Real-Time quantitative PCR (RTqPCR), which contains primers and probes for the targeted viroid and for plant mRNA as internal RTqPCR positive control (IPC) (Guček et al., 2016; Seigner et al., 2020). However, this best-practice strategy for fresh plant material has limitations when it comes to the analysis of fermentation residues likely because of plant mRNA degradation. Without plant mRNA the common IPC is not functioning; thus, false negative results cannot be ruled out. Therefore, an alternative PCR strategy has been tested by using the ribosomal RNA of methanogenic archaea as IPC. In brief, the fate of the HLVd during fermentation in well-established biogas plant model setup was studied and the RTqPCR strategy was specifically adapted for this experiment as foundations for high-throughput screenings. The results have direct implications for hop producers and biogas plant operators using hop harvest residues as substrate.
Materials and methods
Plant material and fermentation experiment
Samples derived from hop harvest residues in the form of chopped bines were gathered directly after harvesting the cultivar “Adriana” from a commercial farm in the hop growing region Hallertau, Germany, in 2019. The plant material was transported to the University of Hohenheim (Stuttgart, Germany) and some samples have been stored at approximately −80 °C directly for the use as pre-treatment reference (pre), while the remaining material was used as substrate for the Hohenheim biogas yield test (HBT). The HBT is a model setup that reliably reproduces the conditions of a biogas plant (Holliger et al., 2016; Hülsemann et al., 2020; Mittweg et al., 2012). The HBT was conducted at two fermentation temperatures of 40 °C (mesophilic conditions) and 50 °C (thermophilic conditions) according to VDI 4630 (VDI protocol, 2016). The fermentation inoculum was a mixture (1:1) taken from two commercial biogas plants, one with a thermophilic biocenosis (biogas plant near Stuttgart run at ~50 °C) and one with a mesophilic biocenosis adapted to digesting hop plant material (Hallertau BIOGAS plant run at 42.5 °C). The purpose of mixing the inocula was to produce a mixed biocenosis, which can quickly adapt to either mesophilic or thermophilic conditions. Each digester syringe (maximum volume 100 ml) was prepared with 1.1 g of hop chaff (31.7% dry matter) together with 30 g biogas inoculum (4.1% dry matter) to achieve an inoculum to substrate dry matter ratio of greater than 2 according the VDI 4630 protocol. Samples were taken after an incubation of 1 and 12 h and after 1, 5, 30 and 90 days, respectively. Three digestion syringes were analyzed for each sampling event. Each sample was immediately frozen and then stored at approximately −80 °C until further processing. Six syringes were operation at 37 °C for 35 days to determine the specific methane potential according to VDI4630. Dry matter (DM) and organic dry matter (ODM) was determined according to DIN EN 15935.
Temperature experiment
For testing the effect of temperature on viroid particle degradation non-fermented chopped hop bines have been used. Analog to the fermentation experiment 1.1 g of chopped bines have been put into a 50 ml vial containing 30 ml of preheated water (instead of slurry). Two vials have been prepared for each temperature and transferred to a preheated incubator at either 20 °C, 40 °C, 50 °C, 60 °C, or 70 °C (WTB Binder, Tuttlingen, Germany). The samples have been incubated for 1 day, 3 days, or 5 days. Thereafter, the water has been removed and the remaining plant material was shock-frozen and stored at −80 °C until RNA extraction and RTqPCR analysis.
RNA extraction and PCR analysis
For the RNA extraction approximately 100 mg of material was used per analysis. The Monarch Total RNA Miniprep Kit (New England Biolabs, Ipswich, USA), was used as previously described (Hagemann et al., 2021). The RTqPCR reaction was calculated for a template volume of 2.5 μl in a total volume of 25 μl per reaction. Based on previous experience it was decided to work with RNA extract dilution of 1:5 and 1:10, respectively, in order to account for possible inhibitory effects. The RTqPCR reaction was performed with the Bioline SensiFAST™ Probe No-ROX One-Step Kit (Bioline, London, UK) on a Rotor Gene Cycler (Rotor Gene 6000, Qiagen, Hilden, Germany) under conditions as previously described (Seigner et al., 2020). Two different duplex RTqPCR have been performed (Table 1); first all samples have been analyzed with a HLVd-nad5-RTqPCR, which was developed for the detection of HLVd and the nad5-mRNA based IPC in fresh plant material (Botermans et al., 2013; Guček et al., 2016; Seigner et al., 2020). Because the nad5-IPC is not well-suited for the given sample matrix (fermentation residues) the HLVd-Arc-RTqPCR duplex strategy was established. The IPC of the latter strategy targets ribosomal RNA of archaea (Lebuhn et al., 2016). Additionally, the HLVd-nad5-RTqPCR was repeated in an independent laboratory at the Bavarian State Research Center for Agriculture (LfL; Freisung, Germany) strictly following the procedures described in Seigner et al. (2020), i.e. in the LfL laboratory the CBCVd-qPCR-R primer was used instead of the CBCV_R1 primer used in the laboratory of the University of Hohenheim (Stuttgart, Germany) (Table 1).
Testing the effect of the sample matrix on the RTqPCR reaction
To test how the qPCR part of the RTqPCR reaction is inhibited by the sample matrix (slurry) the following analysis has been conducted analog to the work of Seigner et al. (2020). A dilution series was conducted in water over six times the power of 10 (107 to 102 copies per μl) with a synthetic HLVd and CBCVd plasmid. 1 μl of each dilution was tested in three technical repetitions to determine the RTqPCR sensitivity in the absence and presence of RNA extract from slurry. 0.5 μl of a HLVd- and CBCVd-free RNA extract from the slurry have been added to half of the RTqPCR reactions. The CBCVd and HLVd plasmids have been produced by integrating the viroid sequence NC_003539 and KT600318, respectively, into the multiple cloning site of the plasmid pEX-A128 (Gene Synthesis Service at Eurofins Genomics, Ebersberg, Germany). The HLVd accession KT600318 used here as well as for the RNA synthesis contains two sequence variations, which prevents a HLVd-typical secondary structure and is therefore presumably not able to form infectious particles. To test how the reverse transcription part of the RTqPCR reaction is inhibited by slurry RNA extract and to estimate the threshold for detection viroid RNA, a HLVd RNA has been synthesized. The template for the RNA synthesis was a T7 promotor followed by the DNA sequence analog to the HLVd accession KT600318, which resulted in a 275 bp long DNA fragment (Gene Synthesis Service at Eurofins Genomics, Ebersberg, Germany). The DNA fragment was delivered lyophilized thus it was resuspended in 50 μl of deionized sterile water, which resulted in a concentration of 6 ng DNA per μl (too little for RNA synthesis). The DNA fragment was used as template for a PCR necessary to increase the amount of DNA to >1 μg as required for a successful RNA synthesis. The PCR was performed with the Q5 High-Fidelity DNA Polymerase set (New England Biolabs, Ipswich, USA) in 50 μl reactions with a standard T7-primer and the newly designed primer HLVd_KT600318_R (Table 1). The resulting amplicon has been purified with the Monarch PCR and DNA Cleanup Kit (New England Biolabs, Ipswich, USA). 1 μg of DNA was used as template for the HiScribe T7 from the according HiScribe T7 Quick High Yield RNA Synthesis Kit (New England Biolabs, Ipswich, USA). The RNA synthesis reaction was incubated at 37 °C overnight. Subsequently, after degrading all DNA by a DNaseI treatment, the RNA was purified following the protocol of the Monarch RNA Cleanup Kit (New England Biolabs, Ipswich, USA). The resulting RNA extract was quantified in four repetitive measurements (Nanodrop 2000 spectrophotometer; Thermo Fisher, Waltham, USA). This resulted into a final concentration of 3.414 μg μl−1, which is equal to 2.325 × 1013 copies per μl considering the following formula with ssRNA mass of 1 μl (3.414 × 10−6 g), ssRNA length (275 nt), average RNA nucleotide mass (321.47 g mol−1 nt−1), hydroxyl- and protonated ends mass accounting for the molecules’ linearity (18.02 g mol−1), and the Avagadro’s number (6.022 × 1023 molecules mol−1):
-
$$ RNA\ copies=\left(\frac{ssRNA\ mass}{ssRNA\ lenght\times nucleotide\ mass+ hydroxyl- ends\ mass}\right)\times {\mathrm{Avagadro}}^{\prime}\mathrm{s}\ \mathrm{number} $$
The extract was diluted to achieve a final concentration of 1 × 1011 copies per μl, which was used for a dilution series (107–102 copies per μl). To test how the reverse transcription is inhibited by slurry RNA extract, the RTqPCR of the RNA dilution series was repeated twice using 1 μl of diluted synthesized HLVd RNA as template, either with or without a 0.5 μl of a HLVd-free slurry RNA extract.
RT-PCR and sequencing
To verify the results of the HLVd-nad5-RTqPCR a HLVd-RT-PCR has been performed at the laboratory of the LfL with the RNAs extracted at the University of Hohenheim (Table 1). The RT-PCR was conducted with the SuperScript™ III One-Step RT-PCR System (Thermo Fischer Scientific, Waltham, USA) with HLVd full length primers (Eastwell & Nelson, 2007). The resulting amplicons were sequenced each with a forward and reverse primer by Sanger sequencing (Sequiserve GmbH, Vaterstetten, Germany) for one representative sample per sample date and for 40 °C and 50 °C samples from day 90. To verify the specificity of the RTqPCR representative amplicons of the HLVd-nad-RTqPCR and HLVd-Arc-RTqPCR have been purified and sequenced each with a forward and reverse primer by Sanger sequencing (Eurofins Genomics, Ebersberg, Germany). The resulting sequences have been assembled and blasted against the NCBI nucleotide database (Geneious 11.1.5 software package, Auckland, New Zealand).
Computational analysis and statistics
Based on the preceding results of the blast analysis the HLVd accessions KT600318 and NC_003611 has been used as input for the secondary structure analysis. This analysis showed that the KT600318 contain sequence variations that prevent HLVd-typic folding patterns and only NC_003611 has been used for predicting secondary structures at different temperatures (20–70 °C). The thermodynamically optimized model prediction was done following the energy model of Andronescu et al. (2007) (Geneious 11.1.5 software package, Auckland, New Zealand). The Cq-values of the RTqPCR (Rotor Gene 6000 Series Software 1.7, Qiagen, Hilden, Germany) were statistically analyzed by pairwise comparison of the means at a probability level of p ≤ 0.01 and Tukey’s honest significant difference (HSD) (R version 3.6.1). The Model assumptions analysis of variance was checked by examining the residual plots and the presented results are the estimates averaged of the levels of the factor dilution (1:5 and 1:10), since it was non-significant.
Results and discussion
RTqPCR adoption to viroid analysis in biogas fermentation residues
The CBCVd is a highly aggressive pathogen for hop that requires working in a quarantine station approved by the local plant protection service (at least in Germany). Therefore, instead of working with CBCVd the ubiquitous HLVd has been used as viroid model. However, to support CBCVd monitoring activities, the potential PCR inhibiting effect of biogas slurry has been evaluated for both viroids comparing RTqPCR performance with or without additional RNA extract from viroid-free slurry. In the first experiment aiming at studying the effect of slurry RNA extract on the qPCR part of the RTqPCR, CBCVd or HLVd containing plasmids (DNA) have been used as template. The results show that there is no inhibitory effect on the qPCR reaction (Fig. 1). In most cases there was no detectable amplicon in the none-template-control (ntc), however, in some samples the ntc resulted in a curve at high Cq-vales. The lowest ntc-based Cq-value is presented as ntc-level in the RTqPCR figures (Figs. 1 and 2). It is important to note that sequencing of the ntc-amplicons did not result in viroid sequences and are artifacts from the qPCR reaction. The qPCR sensitivity of both viroids reactions is highly similar and shows on average Cq-value differences of 3.3 cycles by one power of ten up to a concentration of 102 plasmid copies per RTqPCR reaction, which indicates an optimal qPCR reaction. The qPCR also produced amplicons with lower copy numbers, but results were not stable. In detail, RTqPCR reactions starting from 102 copies per qPCR reaction showed high standard deviation and the Cq-values were close to the ntc-level. Consequently, the limit of reliable viroid detection of either HLVd or CBCVd viroid fragment is above 102 copies per qPCR reaction.
Result of a RTqPCR dilution analysis of HLVd (a) and CBCVd (b) containing plasmids with addition of either water (black square) or viroid-free slurry (grey rhombus). The abscissa show the calculated number of plasmids per PCR reaction μl and the ordinates show the RTqPCR results as Cq-values with the ntc-level indicated. The same letters indicate no statistical difference (p > 0.01, Tukey HSD test)
Result of a RTqPCR dilution analysis of synthetic HLVd RNA with addition of either water (black square) or viroid-free slurry (grey rhombus). The abscissa shows the calculated number of RNA fragments per μl and the ordinate shows the RTqPCR results as Cq-values with the ntc-level indicated. The same letters indicate no statistical difference (p > 0.01, Tukey HSD test)
The HLVd and CBCVd qPCR tests using DNA as template show that performance of detection of both viroids is highly comparable, supporting that HLVd detection can be used as model qPCR for the CBCVd qPCR analysis. For this reason and to reduce the risk of producing infectious CBCVd-RNA, the next experiment, which was conducted to study the possible inhibiting effect of slurry RNA on the overall RTqPCR process was conducted only with synthetic HLVd-RNA of a presumably non-functional HLVd accession, which has critical mutations at the central conserved region (CCR). The result of the RTqPCR evaluation based on synthetic RNA are in good agreement with the results of the qPCR evaluation based on DNA (plasmid); there are no statistical differences between the samples with additional slurry RNA extract and those with only water, the Cq-values increase approximately 3.3 per one power of 10 (Fig. 2). However, the comparison of the Cq-values from the RTqPCR starting from DNA (plasmid) and RNA (synthetic HLVd) show a discrepancy of a Cq-value of only 1.8 (Fig. 2). 1.8 is equal to a 51% lower efficiency of the RNA-based RTqPCR reaction compared to the DNA-based reaction (base-3.3 logarithm), when comparing the same initial copy number of viroid sequences. Generally, the RT is a process which is sensitive to inhibitory effects and can be considered as the bottleneck of RTqPCR reactions; therefore, especially at low concentrations of templates RNA levels are underestimated (Chandler et al., 1998). At a level of 103 copies per reaction the Cq-values are close to the ntc-level, thus 104 copies per reaction are considered as threshold for reliably measurements.
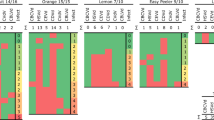
Hop slurry seems to be a difficult sample matrix because the plant material is already partly degraded by the temperature, low pH-value, organic acids, and microbial activities. In a first attempt the well-established nad5-transcript has been tested as IPC (Guček et al., 2019; Seigner et al., 2020). The nad5-IPC worked reliably for the chopped bines, which acted as pre-treatment reference (Fig. 3a). However, as soon as the fermentation started the level of nad5 declined close to the ntc-level already after 12 h of fermentation and thereafter. Therefore, in search for an alternative IPC the methanogen archaea have been targeted, because they are the predominant microorganisms of typical biogas plant biocenosis (Lebuhn et al., 2016). The results show that the targeted 16S-RNA of archaea is only present in limited concentration in the chopped hop bines but they are highly present in all fermentations samples (Fig. 3b). Because the archaea RNA works well as IPC the HLVd levels can be meaningfully interpreted (Fig. 3c).
RTqPCR results of the internal PCR control (IPC) for (a) nad5-transcript, (b) the archea-specific 16S transcript and (C) of the hop latent viroid (HLVd). The fermentation conditions were mesophilic (40 °C; black square) or thermophilic (50 °C; grey rhombus). The abscissa show the incubation time and the ordinates show the RTqPCR results as Cq-values with the ntc-level indicated. The same letters indicate no statistical difference (p > 0.01, Tukey HSD test)
Effect of the biogas fermentation process on HLVd degradation
The HLVd levels seem to decline during the fermentation process. Here it is important to note that the first increase of the Cq-values between the pre-treatment (bines) and 1 h (slurry) is a result from mixing the bines with the biogas-inoculum and is not due to viroid degradation. The further increase of Cq-values could be attributed to degradation reactions as observed at 30 days for the mesophilic conditions (40 °C) and already at 5 days for the thermophilic conditions (50 °C); finally, at 90 days Cq-values reach a level similar to the ntc-level. However, Sanger sequencing of representative full-length RT-PCR amplicons per sample day showed that, albeit at low levels, there are still full-length viroid fragments detectable in the slurry up to 90 days and at 50 °C. Further, the HLVd analysis with the HLVd-nad-RTqPCR of both laboratories support the analysis with the HLVd-arc-RTqPCR that the HLVd concentration significantly decreased after 30 days (40 °C) and 5 days (50 °C). Experiments with composting of viroid-infected tomato residues showed that another member of the Pospiviroidae - the PSTVd drops below detection limit within a week at temperatures of 50 °C, while it took 4 weeks under ambient temperature conditions (Kerins et al., 2018). Certainly, not only temperature and duration time are crucial factors for sanitization. In an earlier study on HLVd infected hop bines it could be shown that even after 16 weeks of ensiling the level of HLVd remained high (Hagemann et al., 2021). However, the study also described that hop ensiling is difficult due to the woody material and the low water content of the bines, which does not allow proper anaerobic ensiling. In addition to the physical parameters also microbial activity can be an important factor for the degradation of plant material and viruses as it is assumed for the thermostable tobamoviruses (Noble & Roberts, 2004). Considering the structural similarities of the HLVd and PSTVd like the rod-like shape and a similar length it is likely that both viroids have a similar level of resistance against physical influences which occur during composting, i.e. microbial activity, higher than ambient temperature and time. To identify the key parameters of the fermentation which might enhance viroid degradation, key process parameters have been measured (Fig. 4). The specific methane potential of hop was measured to 182 l/kgODM, the DM to 29.4% and ODM to 92.6 %DM. Other key parameters of the batch-fermentation process are presented in Fig. 4, showing that there is a temperature independent increase of propionic acid and acetic acid until day 1 and day 5, respectively (Fig. 4 A and B). Both acids appear as intermediate products during the biogas process. This corresponds to a decrease of the pH-value until day 1 (Fig. 4c), thereafter, the level of organic acids decreases to a low level where it remains. The pH is mostly around a value of pH 8 throughout the measure dates, except a pH-value of 8.7 only at the thermophilic conditions after 90 days of fermentation. Generally a pH range between 7.0 and 8.5 indicates an optimal biogas plant fermentation (Holliger et al., 2016). Furthermore, the rapid decrease of plant material (rapid decrease of nad5-RNA and volatile fatty acids) along with constantly high abundance of archaea support the assumption that the biogas process was operating well. No inhibition was reported at both temperatures.
Effect of temperature on HLVd degradation under non-fermenting conditions
To further examine the temperature effect on viroid degradation an additional experiment was conducted under non-fermenting conditions (Fig. 5). Here incubation at room temperature at 20 °C, was compared to fermentation temperatures 40 °C and 50 °C as well as higher temperatures of 60 °C and 70 °C. Temperatures higher than 70 °C have not been considered since the according energy requirement might be too high (expensive) to have practical implications for potential pretreatments. The results show that at 20 °C there is no difference in viroid levels between any sample day (1, 3, and 5) (Fig. 5) indicating that there is no viroid degradation at 20 °C at least up to the final day of the experiment. Generally, as the temperature increases also the Cq-values increase, which reflects less detectable viroid in the sample. In detail, after 1 day of incubation only the samples kept at 70 °C showed Cq-values significantly different from the others (Fig. 5). After 3 days Cq-values for 50 °C and 60 °C increased significantly when compared to 20 °C. After 5 days of incubation Cq-values of the samples incubated at 70 °C reached the ntc-level, while Cq-values for 50 °C and 60 °C remained significantly different from 20 °C. Such high temperatures might not have practical implications for the biogas fermentation but a short heat treatment at 70 °C significantly reduces the viroid load and thus could be implemented as pretreatment. The pasteurization at 70 °C for 1 h is a necessary minimum requirement for treating food waste prior to the biogas process, if the residues should be used as agricultural fertilizer in the EU (EC regulation 1774/2002, 3 October 2002). Furthermore, the finding that the temperature of 70 °C reduced the HLVd concentration by four decimal powers already within 1 day agrees with the PSTVd composting experiments of Kerins et al. (2018); the authors shown that in moist compost at 65 °C complete viroid degradation could be observed 2 days after incubation, while it took a week at 50 °C. This raises the question about the principal differences between an incubation temperature of about 70 °C (or 65 °C for the PSTVd) and lower temperatures of about 50 °C.
Barplot of the RTqPCR results from the experiment of measuring the effect of temperature on the HLVd. The abscissa shows the incubation time and the ordinate shows the RTqPCR results as Cq-values with the ntc-level indicated. The same letters indicate no statistical difference (p > 0.01, Tukey HSD test)
Secondary structural prediction HLVd and CBCVd at high temperatures
Steger and Riesner (2018) summarize that the rod-like structure of pospiviroids starts dissociating with increasing temperatures. Indeed, based on the calculations of this study the viroids destabilize as the temperature increases (Table 2), which is reflected by lower values for the minimal free energy as calculated based on the model of Andronescu et al. (2007). CBCVd and HLVd both show differences in their left terminal loop region already at 40 °C compared to 20 °C. The structural differences at 40 °C, 50 °C and 60 °C are rather small, however, profound differences occur when increasing the temperature from 60 to 70 °C for both viroids. Further increasing the temperature until the minimal free energy equals zero, which is equal to a complete dissociation of the rod-like structure, occurs at critical temperatures of 71 °C for the HLVd (NC003611), 75 °C for the CBCVd (NC_003539), and 78 °C for PSTVd (AJ583449). For PSTVd melting analysis have shown that the PSTVd is reshaping at 68 °C to form an almost fully dissociated metastructure in vitro (Henco et al., 1979). These type of (almost) complete dissociation close to 70 °C might increases viroid decay by making the RNA accessible to RNA silencing or to RNase-mediated endonucleolytic cleavage. Especially, RNases of extremophilic bacteria and archaea are resistant to harsh environmental conditions; for example, the RNase from Thermus aquaticus still shows 20% of their maximal activity at 75 °C (Brown et al., 1993). Furthermore, RNases of methanogenic archaea have a temperature optimum at 60 °C and are still active at 70 °C (Levy et al., 2011). However, the details about the main processes driving viroid turnover are not yet fully understood (Minoia et al., 2015).
Conclusion
Based on the presented results it can be concluded that the HLVd and presumably other pospiviroids are difficult to eliminate even with the biogas fermentation process. HLVd was still detectable after 90 days of thermophilic fermentation in a well-functioning biogas plant model system. However, the biogas process can contribute to viroid reduction, as for example the HLVd was significantly reduced after 30 days at 40 °C or after 5 days at 50 °C, respectively. Further, our experiments show that a pretreatment of HLVd-infected hop harvest residues with temperatures of 70 °C can reduce the viroid level within only 1 day. Therefore, we suggest a combinatory process with a heat pre-treatment followed by thermophilic biogas fermentation. This two-step process will produce not just renewable energy but might also deliver a safe bio-fertilizer for plant production. However, the methane potential of hop is only 53% of the methane potential of maize (340 l/kgODM) but hop harvest residues are low-cost by-product. Therefore, biogas plant operators properly need to get paid for taking potentially contaminated substrate to get a sustainable business model that can be part of a viroid decontamination strategy.
References
Andronescu, M., Condon, A., Hoos, H. H., Mathews, D. H., & Murphy, K. P. (2007). Efficient parameter estimation for RNA secondary structure prediction. Bioinformatics, 23, i19–i28. https://doi.org/10.1093/bioinformatics/btm223.
Bar-Joseph, M. (1993). Citrus viroids and citrus dwarfing in Israel. Acta Horticulturae, 349, 271–276. https://doi.org/10.17660/ActaHortic.1993.349.45.
Bektaş, A., Hardwick, K. M., Waterman, K., & Kristof, J. (2019). Occurrence of Hop latent viroid in Cannabis sativa with symptoms of cannabis stunting disease in California. Plant Disease, 103(10), 2699. https://doi.org/10.1094/PDIS-03-19-0459-PDN.
Botermans, M., van de Vossenberg, B. T. L. H., Verhoeven, J. T. J., Roenhorst, J. W., Hooftman, M., Dekter, R., & Meekes, E. T. M. (2013). Development and validation of a real-time RT-PCR assay for generic detection of pospiviroids. Journal of Virology Methods, 187, 43–50. https://doi.org/10.1016/j.jviromet.2012.09.004.
Brown, J. W., Haas, E. S., & Pace, N. R. (1993). Characterization of ribonuclease P RNAs from thermophilic bacteria. Nucleic Acids Research, 21(3), 671–679. https://doi.org/10.1093/nar/21.3.671.
Chandler, D. P., Wagnon, C. A., & Bolton, H. (1998). Reverse transcriptase (RT) inhibition of PCR at low concentrations of template and its implications for quantitative RT-PCR. Applied Environmental Microbiology, 64, 669–677. https://doi.org/10.1128/aem.64.2.669-677.1998.
Diener, T. O. (1972). Is the scrapie agent a viroid? Nature New Biology, 235, 218–219. https://doi.org/10.1038/newbio235218a0.
Di Serio, F., Flores, R., Verhoeven, J. Th J., Li, S.-F., Pallás, V., Randles, J. W., Sano, T., Vidalakis, G., & Owens, R. A. (2014). Current status of viroid taxonomy. Archives of Virology, 159(12), 3467–3478. https://doi.org/10.1007/s00705-014-2200-6.
Eastwell, K. C., & Nelson, M. E. (2007). Occurrence of viroids in commercial hop (Humulus lupulus L.) production areas of Washington state. Plant Health Progress, 8(4), 1–8. https://doi.org/10.1094/PHP-2007-1127-01-RS.
Guček, T., Štajner, N., Jakše, J., & Javornik, B. (2016). Optimization of detection of Hop latent viroid (HLVd) with real time RT-PCR. Hop Bulletin, 23, 27–40.
Guček, T., Jakše, J., Matoušek, J., & Radišek, S. (2019). One-step multiplex RT-PCR for simultaneous detection of four viroids from hop (Humulus lupulus L.). European Journal of Plant Pathology, 154, 273–286. https://doi.org/10.1007/s10658-018-01654-2.
Hagemann, M. H., Born, U., Sprich, E., Seigner, L., Oechsner, H., Hülsemann, B., Steinbrenner, J., Wünsche, J.N., & Lehmair, E. (2021). Fate of the hop latent viroid during ensiling of hop harvest residues. In Hagemann, M. H. & Wünsche, J. N., (Eds), International Humulus Symposium, Acta Horticulturae. International Society for Horticultural Science, Leuven.
Henco, K., Sänger, H. L., & Riesner, D. (1979). Fine structure melting of viroids as studied by kinetic methods. Nucleic Acids Research, 6, 3041–3059. https://doi.org/10.1093/nar/6.9.3041.
Holliger, C., Alves, M., Andrade, D., Angelidaki, I., Astals, S., Baier, U., Bougrier, C., Buffière, P., Carballa, M., De Wilde, V., Ebertseder, F., Fernández, B., Ficara, E., Fotidis, I., Frigon, J. C., De Laclos, H. F., Ghasimi, D. S. M., Hack, G., Hartel, M., et al. (2016). Towards a standardization of biomethane potential tests. Water Science and Technology, 74, 2515–2522. https://doi.org/10.2166/wst.2016.336.
Hülsemann, B., Zhou, L., Merkle, W., Hassa, J., Müller, J., & Oechsner, H. (2020). Biomethane potential test: Influence of inoculum and the digestion system. Applied Science, 10(7), 2589. https://doi.org/10.3390/app10072589.
Jakse, J., Radisek, S., Pokorn, T., Matousek, J., & Javornik, B. (2015). Deep-sequencing revealed Citrus bark cracking viroid (CBCVd) as a highly aggressive pathogen on hop. Plant Pathology, 64, 831–842. https://doi.org/10.1111/ppa.12325.
Kerins, G., Blackburn, J., Nixon, T., Daly, M., Conyers, C., Pietravalle, S., Noble, R., & Henry, C. M. (2018). Composting to sanitize plant-based waste infected with organisms of plant health importance. Plant Pathology, 67, 411–417. https://doi.org/10.1111/ppa.12729.
Lebuhn, M., Derenkó, J., Rademacher, A., Helbig, S., Munk, B., Pechtl, A., Stolze, Y., Prowe, S., Schwarz, W. H., Schlüter, A., Liebl, W., & Klocke, M. (2016). DNA and RNA extraction and quantitative real-time PCR-based assays for biogas biocenoses in an interlaboratory comparison. Bioengineering, 3(1), 7. https://doi.org/10.3390/bioengineering3010007.
Levy, S., Portnoy, V., Admon, J., & Schuster, G. (2011). Distinct activities of several RNase J proteins in methanogenic archaea. RNA Biology, 8(6), 1073–1083. https://doi.org/10.4161/rna.8.6.16604.
Matsushita, Y. (2013). Chrysanthemum stunt viroid. Japan International Research Center for Agricultural Sciences, 47(3), 237–242. https://doi.org/10.6090/jarq.47.237.
Mehle, N., Gutiérrez-Aguirre, I., Prezelj, N., Delić, D., Vidic, U., & Ravnikar, M. (2014). Survival and transmission of potato virus Y, Pepino mosaic virus, and potato spindle tuber viroid in water. Applied Environmental Microbiology, 80, 1455–1462. https://doi.org/10.1128/AEM.03349-13.
Minoia, S., Navarro, B., Delgado, S., Di Serio, F., & Flores, R. (2015). Viroid RNA turnover: Characterization of the subgenomic RNAs of potato spindle tuber viroid accumulating in infected tissues provides insights into decay pathways operating in vivo. Nucleic Acids Research, 43, 2313–2325. https://doi.org/10.1093/nar/gkv034.
Mittweg, G., Oechsner, H., Hahn, V., Lemmer, A., & Reinhardt-Hanisch, A. (2012). Repeatability of a laboratory batch method to determine the specific biogas and methane yields. Engineering in Life Sciences, 12, 270–278. https://doi.org/10.1002/elsc.201100181.
Noble, R., & Roberts, S. J. (2004). Eradication of plant pathogens and nematodes during composting: A review. Plant Pathology, 53, 548–568. https://doi.org/10.1111/j.0032-0862.2004.01059.x.
Oechsner, H. (2017). Putting the discussion on biogas on an objective footing is absolutely essential. Landtechnik, 72, 263–264. https://doi.org/10.15150/lt.2017.3172.
Radisek, S. (2017). Pest risk analysis for Citrus bark cracking viroid (CBCVd). European Plant Protection Organisation. Retrieved August 16, 2021, from https://gd.eppo.int/download/doc/1263_pra_exp_CBCVd.pdf.
Seigner, L., Kappen, M., Huber, C., Kistler, M., & Köhler, D. (2008). First trials for transmission of potato spindle tuber viroid from ornamental Solanaceae to tomato using RT-PCR and an mRNA based internal positive control for detection. Journal of Plant Diseases and Protection, 115, 97–101. https://doi.org/10.1007/BF03356246.
Seigner, L., Liebrecht, M., Keckel, L., Einberger, K., & Absmeier, C. (2020). Real-time RT-PCR detection of Citrus bark cracking viroid (CBCVd) in hops including an mRNA-based internal positive control. Journal of Plant Diseases and Protection, 127, 763–767. https://doi.org/10.1007/s41348-020-00317-x.
Škorić, D. (2017). Viroid biology. In: Hadidi, A (Ed) Viroids and Satellites (pp 53–61). Elsevier Inc., Amsterdam. https://doi.org/10.1016/B978-0-12-801498-1.00005-X.
Steger, G., & Riesner, D. (2018). Viroid research and its significance for RNA technology and basic biochemistry. Nucleic Acids Research, 46(20), 10563–10576. https://doi.org/10.1093/nar/gky903.
Takahashi, T., & Yaguchi, S. (1985). Strategies for preventing mechanical transmission of hop stunt viroid. Journal of Plant Diseases and Protection, 92(2), 132–137.
VDI protocol (2016). VDI 4630. Fermentation of organic materials – characterisation of the substrate, sampling, collection of material data, data, fermentation tests. Verlag des Vereins Deutscher Ingenieure, Düsseldorf.
Verhoeven, J. T. J., Hüner, L., Marn, M. V., Plesko, I. M., & Roenhorst, J. W. (2010). Mechanical transmission of potato spindle tuber viroid between plants of Brugmansia suaveoles, Solanum jasminoides and potatoes and tomatoes. European Journal of Plant Pathology, 128, 417–421. https://doi.org/10.1007/s10658-010-9675-0.
Vidalakis, G., Pagliaccia, D., Bash, J. A., Afunian, M., & Semancik, J. S. (2011). Citrus dwarfing viroid: Effects on tree size and scion performance specific to Poncirus trifoliata rootstock for high-density planting. Annals of Applied Biology, 158(2), 204–217. https://doi.org/10.1111/j.1744-7348.2010.00454.x.
Warren, J. G., Mercado, J., & Grace, D. (2019). Occurrence of hop latent viroid causing disease in Cannabis sativa in California. Plant Disease, 103(10), 2699–2699. https://doi.org/10.1094/PDIS-03-19-0530-PDN.
Acknowledgements
We thank Prof. Dr. Jens N. Wünsche † for contributed to the initiation and planning of this work as well as for his guidance along the way.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
There is no conflict of interest.
Data availability
Not applicable.
Code availability
Not applicable.
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Hagemann, M.H., Born, U., Sprich, E. et al. Degradation of hop latent viroid during anaerobic digestion of infected hop harvest residues. Eur J Plant Pathol 161, 579–591 (2021). https://doi.org/10.1007/s10658-021-02344-2
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10658-021-02344-2