Abstract
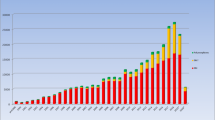
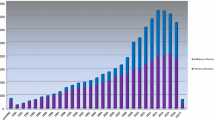
This article provides a historical overview of the online database (www.insight-group.org/mutations) maintained by the International Society for Gastrointestinal Hereditary Tumours. The focus is on the mismatch repair genes which are mutated in Lynch Syndrome. APC, MUTYH and other genes are also an important part of the database, but are not covered here. Over time, as the understanding of the genetics of Lynch Syndrome increased, databases were created to centralise and share the variants which were being detected in ever greater numbers. These databases were eventually merged into the InSiGHT database, a comprehensive repository of gene variant and disease phenotype information, serving as a starting point for important endeavours including variant interpretation, research, diagnostics and enhanced global collection. Pivotal to its success has been the collaborative spirit in which it has been developed, its association with the Human Variome Project, the appointment of a full time curator and its governance stemming from the well established organizational structure of InSiGHT.
Similar content being viewed by others
References
Fokkema IF, Taschner PE, Schaafsma GC et al (2011) LOVD v. 2.0: the next generation in gene variant databases. Hum Mutat 32:557–563
Peltomäki P, Vasen H (1997) Mutations predisposing to hereditary nonpolyposis colorectal cancer: database and results of a collaborative study. The International Collaborative Group on Hereditary Nonpolyposis Colorectal Cancer. Gastroenterology 113:1146–1158
Peltomäki P, Vasen H (2004) Mutations associated with HNPCC predisposition—update of ICG-HNPCC/INSiGHT mutation database. Dis Markers 20:269–276
Woods MO, Williams P, Careen A et al (2007) A new variant database for mismatch repair genes associated with Lynch syndrome. Hum Mutat 28:669–673
Ou J, Niessen RC, Vonk J et al (2008) A database to support the interpretation of human mismatch repair gene variants. Hum Mutat 29:1337–1341
Kaput J, Cotton RG, Hardman L et al (2009) Planning the Human Variome Project: the Spain report. Hum Mutat 30:496–510
Plon SE, Eccles DM, Easton D et al (2008) Sequence variant classification and reporting: recommendations for improving the interpretation of cancer susceptibility genetic test results. Hum Mutat 29:1282–1291
Giardine B, Borg J, Higgs DR et al (2011) Systematic documentation and analysis of human genetic variation in hemoglobinopathies using the microattribution approach. Nat Genet 43(4):295–301
Acknowledgments
The interpretation committee is operationally funded through the Cancer Council of Victoria (Australia), and the curation generously funded by the George Hicks Foundation in Melbourne Australia (to September 2012) and then The Royal Melbourne Hospital Foundation.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Plazzer, J.P., Sijmons, R.H., Woods, M.O. et al. The InSiGHT database: utilizing 100 years of insights into Lynch Syndrome. Familial Cancer 12, 175–180 (2013). https://doi.org/10.1007/s10689-013-9616-0
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10689-013-9616-0