Abstract
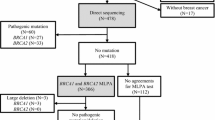
Large genomic rearrangements (LGRs) account for at least 10 % of the mutations in BRCA1 and 5 % of BRCA2 mutations in outbred hereditary breast and ovarian cancer (HBOC) families. Data from some series suggest LGRs represent particularly penetrant mutations. 1,034 index cases from HBOC families underwent comprehensive BRCA1 and BRCA2 mutation testing, including screening for LGRs. The personal and family history of 254 identified mutation carriers were compared based on mutation type. Thirty-six LGRs were detected; 32/122 (26 %) BRCA1 and 4/132 (3 %) BRCA2 mutations. High risk features (bilateral breast cancer, diagnosis <40 years, ovarian cancer, male breast cancer) were more commonly associated with an LGR than a non-LGR mutation (p = 0.008), In families with a BRCA1 LGR the mean age of breast cancer diagnosis was younger than in families with a non-LGR BRCA1 mutation (42.5 vs. 46.1 years, p = 0.007). Across the entire group of mutation positive families the number of relatives affected by breast or ovarian cancer was increased [LGR 3.7 vs. non- LGR 2.8 per family, p value (adjusted for genotype) = 0.047]. Excluding index cases, the odds ratio for breast cancer in BRCA1 families with an LGR was 1.42 (95 % CI 1.24–1.63) and for ovarian cancer 1.66 (95 % CI 1.10–2.49). The increased cancer risk was reflected in significantly higher risk assessments by mutation prediction tools. LGRs are associated with higher cancer risks. If validated, LGRs could be included in cancer risk prediction tools to improve personalised cancer risk prediction estimates and may guide cost-minimising mutation screening strategies in some healthcare settings.


Similar content being viewed by others
Abbreviations
- BMI:
-
Body mass index
- CBC:
-
Contralateral breast cancer
- CIMBA:
-
Consortium of Investigators of the Modifiers of BRCA1/2 Alleles
- DHPLC:
-
Denaturing high performance liquid chromatography
- DNA:
-
Deoxyribonucleic acid
- FCC:
-
Familial Cancer Centre
- HBOC:
-
Hereditary breast and ovarian cancer
- LGR:
-
Large genomic rearrangement
- MLPA:
-
Multiplex ligation-dependent probe amplification
- OCCR:
-
Ovarian cancer cluster region
- PCR:
-
Polymerase chain reaction
- PTT:
-
Protein truncation test
References
Alsop K, Fereday S, Meldrum C, deFazio A, Emmanuel C, George J, Dobrovic A, Birrer MJ, Webb PM, Stewart C et al (2012) BRCA mutation frequency and patterns of treatment response in BRCA mutation-positive women with ovarian cancer: a report from the Australian Ovarian Cancer Study Group. J Clin Oncol 30(21):2654–2663
Antoniou A, Pharoah PD, Narod S, Risch HA, Eyfjord JE, Hopper JL, Loman N, Olsson H, Johannsson O, Borg A et al (2003) Average risks of breast and ovarian cancer associated with BRCA1 or BRCA2 mutations detected in case Series unselected for family history: a combined analysis of 22 studies. Am J Hum Genet 72(5):1117–1130
Montagna M, Dalla Palma M, Menin C, Agata S, De Nicolo A, Chieco-Bianchi L, D’Andrea E (2003) Genomic rearrangements account for more than one-third of the BRCA1 mutations in northern Italian breast/ovarian cancer families. Hum Mol Genet 12(9):1055–1061
Engert S, Wappenschmidt B, Betz B, Kast K, Kutsche M, Hellebrand H, Goecke TO, Kiechle M, Niederacher D, Schmutzler RK et al (2008) MLPA screening in the BRCA1 gene from 1,506 German hereditary breast cancer cases: novel deletions, frequent involvement of exon 17, and occurrence in single early-onset cases. Hum Mutat 29(7):948–958
Palma MD, Domchek SM, Stopfer J, Erlichman J, Siegfried JD, Tigges-Cardwell J, Mason BA, Rebbeck TR, Nathanson KL (2008) The relative contribution of point mutations and genomic rearrangements in BRCA1 and BRCA2 in high-risk breast cancer families. Cancer Res 68(17):7006–7014
Hansen TO, Jonson L, Albrechtsen A, Andersen MK, Ejlertsen B, Nielsen FC (2009) Large BRCA1 and BRCA2 genomic rearrangements in Danish high risk breast-ovarian cancer families. Breast Cancer Res Treat 115(2):315–323
Pylkas K, Erkko H, Nikkila J, Solyom S, Winqvist R (2008) Analysis of large deletions in BRCA1, BRCA2 and PALB2 genes in Finnish breast and ovarian cancer families. BMC Cancer 8:146
del Valle J, Feliubadalo L, Nadal M, Teule A, Miro R, Cuesta R, Tornero E, Menendez M, Darder E, Brunet J et al (2010) Identification and comprehensive characterization of large genomic rearrangements in the BRCA1 and BRCA2 genes. Breast Cancer Res Treat 122(3):733–743
Vasickova P, Machackova E, Lukesova M, Damborsky J, Horky O, Pavlu H, Kuklova J, Kosinova V, Navratilova M, Foretova L (2007) High occurrence of BRCA1 intragenic rearrangements in hereditary breast and ovarian cancer syndrome in the Czech Republic. BMC Med Genet 8:32
Ruiz de Garibay G, Gutierrez-Enriquez S, Garre P, Bonache S, Romero A, Palomo L, Sanchez de Abajo A, Benitez J, Balmana J, Perez-Segura P et al (2012) Characterization of four novel BRCA2 large genomic rearrangements in Spanish breast/ovarian cancer families: review of the literature, and reevaluation of the genetic mechanisms involved in their origin. Breast Cancer Res Treat 133(1):273–283
Kim H, Choi DH (2013) Distribution of and mutations in Asian patients with breast cancer. J Breast Cancer 16(4):357–365
Kang P, Mariapun S, Phuah SY, Lim LS, Liu J, Yoon SY, Thong MK, Mohd Taib NA, Yip CH, Teo SH (2012) Large BRCA1 and BRCA2 genomic rearrangements in Malaysian high risk breast-ovarian cancer families. Breast Cancer Res Treat 124(2):579–584
Woodward AM, Davis TA, Silva AG, Kirk JA, Leary JA, kConFab Investigators (2005) Large genomic rearrangements of both BRCA2 and BRCA1 are a feature of the inherited breast/ovarian cancer phenotype in selected families. J Med Genet 42(5):e31
Berry DA, Iversen ES Jr, Gudbjartsson DF, Hiller EH, Garber JE, Peshkin BN, Lerman C, Watson P, Lynch HT, Hilsenbeck SG et al (2002) BRCAPRO validation, sensitivity of genetic testing of BRCA1/BRCA2, and prevalence of other breast cancer susceptibility genes. J Clin Oncol 20(11):2701–2712
Chen S, Wang W, Broman KW, Katki HA, Parmigiani G (2004) BayesMendel: an R environment for Mendelian risk prediction. Statistical Appl Genet Mol Biol 3:Article21
James PA, Doherty R, Harris M, Mukesh BN, Milner A, Young MA, Scott C (2006) Optimal selection of individuals for BRCA mutation testing: a comparison of available methods. J Clin Oncol 24(4):707–715
Evans DG, Eccles DM, Rahman N, Young K, Bulman M, Amir E, Shenton A, Howell A, Lalloo F (2004) A new scoring system for the chances of identifying a BRCA1/2 mutation outperforms existing models including BRCAPRO. J Med Genet 41(6):474–480
Domchek SM, Friebel TM, Singer CF, Evans DG, Lynch HT, Isaacs C, Garber JE, Neuhausen SL, Matloff E, Eeles R et al (2010) Association of risk-reducing surgery in BRCA1 or BRCA2 mutation carriers with cancer risk and mortality. JAMA 304(9):967–975
Chen S, Parmigiani G (2007) Meta-analysis of BRCA1 and BRCA2 penetrance. J Clin Oncol 25(11):1329–1333
Petrucelli N, Daly MB, Feldman GL (2010) Hereditary breast and ovarian cancer due to mutations in BRCA1 and BRCA2. Genet Med 12(5):245–259
Chenevix-Trench G, Milne RL, Antoniou AC, Couch FJ, Easton DF, Goldgar DE (2007) Cimba: an international initiative to identify genetic modifiers of cancer risk in BRCA1 and BRCA2 mutation carriers: the Consortium of Investigators of Modifiers of BRCA1 and BRCA2 (CIMBA). Breast Cancer Res 9(2):104
Moorman PG, Iversen ES, Marcom PK, Marks JR, Wang F, Kathleen Cuningham Consortium for Research into Familial Breast Cancer, Lee E, Ursin G, Rebbeck TR, Domchek SM et al (2010) Evaluation of established breast cancer risk factors as modifiers of BRCA1 or BRCA2: a multi-center case-only analysis. Breast Cancer Res Treat 124(2):441–451
Lubinski J, Phelan CM, Ghadirian P, Lynch HT, Garber J, Weber B, Tung N, Horsman D, Isaacs C, Monteiro AN et al (2004) Cancer variation associated with the position of the mutation in the BRCA2 gene. Fam Cancer 3(1):1–10
Gayther SA, Warren W, Mazoyer S, Russell PA, Harrington PA, Chiano M, Seal S, Hamoudi R, van Rensburg EJ, Dunning AM et al (1995) Germline mutations of the BRCA1 gene in breast and ovarian cancer families provide evidence for a genotype-phenotype correlation. Nat Genet 11(4):428–433
Thompson D, Easton D (2002) Breast cancer linkage C: variation in BRCA1 cancer risks by mutation position. Cancer Epidemiol Biomark Prevent 11(4):329–336
Antoniou AC, Rookus M, Andrieu N, Brohet R, Chang-Claude J, Peock S, Cook M, Evans DG, Eeles R, EMBRACE et al (2009) Reproductive and hormonal factors, and ovarian cancer risk for BRCA1 and BRCA2 mutation carriers: results from the International BRCA1/2 Carrier Cohort Study. Cancer Epidemiol Biomark Prevent 18(2):601–610
Andrieu N, Goldgar DE, Easton DF, Rookus M, Brohet R, Antoniou AC, Peock S, Evans G, Eccles D, Douglas F et al (2006) Pregnancies, breast-feeding, and breast cancer risk in the International BRCA1/2 Carrier Cohort Study (IBCCS). J Natl Cancer Inst 98(8):535–544
Manders P, Pijpe A, Hooning MJ, Kluijt I, Vasen HF, Hoogerbrugge N, van Asperen CJ, Meijers-Heijboer H, Ausems MG, van Os TA et al (2011) Body weight and risk of breast cancer in BRCA1/2 mutation carriers. Breast Cancer Res Treat 126(1):193–202
Mitchell G, Antoniou AC, Warren R, Peock S, Brown J, Davies R, Mattison J, Cook M, Warsi I, Evans DG et al (2006) Mammographic density and breast cancer risk in BRCA1 and BRCA2 mutation carriers. Cancer Res 66(3):1866–1872
Mulligan AM, Couch FJ, Barrowdale D, Domchek SM, Eccles D, Nevanlinna H, Ramus SJ, Robson M, Sherman M, Spurdle AB et al (2011) Common breast cancer susceptibility alleles are associated with tumour subtypes in BRCA1 and BRCA2 mutation carriers: results from the Consortium of Investigators of Modifiers of BRCA1/2. Breast Cancer Res 13(6):R110
Wang X, Pankratz VS, Fredericksen Z, Tarrell R, Karaus M, McGuffog L, Pharaoh PD, Ponder BA, Dunning AM, Peock S et al (2010) Common variants associated with breast cancer in genome-wide association studies are modifiers of breast cancer risk in BRCA1 and BRCA2 mutation carriers. Hum Mol Genet 19(14):2886–2897
Sawyer S, Mitchell G, McKinley J, Chenevix-Trench G, Beesley J, Chen XQ, Bowtell D, Trainer AH, Harris M, Lindeman GJ et al (2012) A role for common genomic variants in the assessment of familial breast cancer. J Clin Oncol 30(35):4330–4336
Spurdle AB, Healey S, Devereau A, Hogervorst FB, Monteiro AN, Nathanson KL, Radice P, Stoppa-Lyonnet D, Tavtigian S, Wappenschmidt B et al (2012) ENIGMA—evidence-based network for the interpretation of germline mutant alleles: an international initiative to evaluate risk and clinical significance associated with sequence variation in BRCA1 and BRCA2 genes. Hum Mutat 33(1):2–7
Acknowledgments
The authors gratefully acknowledge the assistance of Rebecca Driessen and Kylie Shackleton for the Victorian Familial Cancer Clinical Trials Group, along with Megan Rehfisch, Steven Macaskill, Ayiguli Ha, Jamie Thiessen, Elvina Johnston, Chris Michael-Lovatt and the Genetic Counsellors of the Victorian FCCs. This study was supported by funding from the Department of Health, Victoria (Australia). The Victorian Familial Cancer Clinical Trials Group was supported by the Victorian Cancer Agency (EOI09-50). GJL was supported by a NHMRC research fellowship, the Victorian Breast Cancer Research Consortium and State Government Operational Infrastructure Support.
Conflict of interests
The author(s) declare that they have no competing interests.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
James, P.A., Sawyer, S., Boyle, S. et al. Large genomic rearrangements in the familial breast and ovarian cancer gene BRCA1 are associated with an increased frequency of high risk features. Familial Cancer 14, 287–295 (2015). https://doi.org/10.1007/s10689-015-9785-0
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10689-015-9785-0