Abstract
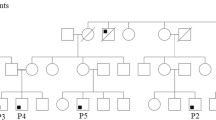
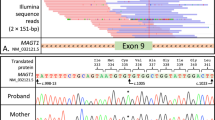
The purpose of this research was to use next generation sequencing to identify mutations in patients with primary immunodeficiency diseases whose pathogenic gene mutations had not been identified. Remarkably, four unrelated patients were found by next generation sequencing to have the same heterozygous mutation in an essential donor splice site of PIK3R1 (NM_181523.2:c.1425 + 1G > A) found in three prior reports. All four had the Hyper IgM syndrome, lymphadenopathy and short stature, and one also had SHORT syndrome. They were investigated with in vitro immune studies, RT-PCR, and immunoblotting studies of the mutation’s effect on mTOR pathway signaling. All patients had very low percentages of memory B cells and class-switched memory B cells and reduced numbers of naïve CD4+ and CD8+ T cells. RT-PCR confirmed the presence of both an abnormal 273 base-pair (bp) size and a normal 399 bp size band in the patient and only the normal band was present in the parents. Following anti-CD40 stimulation, patient’s EBV-B cells displayed higher levels of S6 phosphorylation (mTOR complex 1 dependent event), Akt phosphorylation at serine 473 (mTOR complex 2 dependent event), and Akt phosphorylation at threonine 308 (PI3K/PDK1 dependent event) than controls, suggesting elevated mTOR signaling downstream of CD40. These observations suggest that amino acids 435–474 in PIK3R1 are important for its stability and also its ability to restrain PI3K activity. Deletion of Exon 11 leads to constitutive activation of PI3K signaling. This is the first report of this mutation and immunologic abnormalities in SHORT syndrome.



Similar content being viewed by others
References
Okkenhaug K. Signaling by the phosphoinositide 3-kinase family in immune cells. Annu Rev Immunol. 2013;31:675–704.
Angulo I, Vadas O, Garcon F, Banham-Hall E, Plagnol V, Leahy TR, et al. Phosphoinositide 3-kinase delta gene mutation predisposes to respiratory infection and airway damage. Science. 2013;342(6160):866–71.
Lucas CL, Kuehn HS, Zhao F, Niemela JE, Deenick EK, Palendira U, et al. Dominant-activating germline mutations in the gene encoding the PI(3)K catalytic subunit p110delta result in T cell senescence and human immunodeficiency. Nat Immunol. 2014;15(1):88–97.
Kracker S, Curtis J, Ibrahim MA, Sediva A, Salisbury J, Campr V, et al. Occurrence of B-cell lymphomas in patients with activated phosphoinositide 3-kinase delta syndrome. J Allergy Clin Immunol. 2014;134(1):233–6.
Crank MC, Grossman JK, Moir S, Pittaluga S, Buckner CM, Kardava L, et al. Mutations in PIK3CD can cause hyper IgM syndrome (HIGM) associated with increased cancer susceptibility. J Clin Immunol. 2014;34(3):272–6.
Deau MC, Heurtier L, Frange P, Suarez F, Bole-Feysot C, Nitschke P, et al. A human immunodeficiency caused by mutations in the PIK3R1 gene. J Clin Invest. 2014;124(9):3923–8.
Lucas CL, Zhang Y, Venida A, Wang Y, Hughes J, McElwee J, et al. Heterozygous splice mutation in PIK3R1 causes human immunodeficiency with lymphoproliferation due to dominant activation of PI3K. J Exp Med. 2014;211(13):2537–47.
Lougaris V, Faletra F, Lanzi G, Vozzi D, Marcuzzi A, Valencic E, et al. Altered germinal center reaction and abnormal B cell peripheral maturation in PI3KR1-mutated patients presenting with HIGM-like phenotype. Clin Immunol. 2015;159(1):33–6.
Deau MC, Heurtier L, Frange P, Suarez F, Bole-Feysot C, Nitschke P, et al. A human immunodeficiency caused by mutations in the PIK3R1 gene. J Clin Invest. 2015;125(4):1764–5.
Conley ME, Dobbs AK, Quintana AM, Bosompem A, Wang YD, Coustan-Smith E, et al. Agammaglobulinemia and absent B lineage cells in a patient lacking the p85alpha subunit of PI3K. J Exp Med. 2012;209(3):463–70.
Dyment DA, Smith AC, Alcantara D, Schwartzentruber JA, Basel-Vanagaite L, Curry CJ, et al. Mutations in PIK3R1 cause SHORT syndrome. Am J Hum Genet. 2013;93(1):158–66.
Chudasama KK, Winnay J, Johansson S, Claudi T, Konig R, Haldorsen I, et al. SHORT syndrome with partial lipodystrophy due to impaired phosphatidylinositol 3 kinase signaling. Am J Hum Genet. 2013;93(1):150–7.
Thauvin-Robinet C, Auclair M, Duplomb L, Caron-Debarle M, Avila M, St-Onge J, et al. PIK3R1 mutations cause syndromic insulin resistance with lipoatrophy. Am J Hum Genet. 2013;93(1):141–9.
Schroeder C, Riess A, Bonin M, Bauer P, Riess O, Dobler-Neumann M, et al. PIK3R1 mutations in SHORT syndrome. Clin Genet. 2014;86(3):292–4.
Barcena C, Quesada V, De Sandre-Giovannoli A, Puente DA, Fernandez-Toral J, Sigaudy S, et al. Exome sequencing identifies a novel mutation in PIK3R1 as the cause of SHORT syndrome. BMC Med Genet. 2014;15:51.
Chung BK, Gibson WT. Autosomal dominant PIK3R1 mutations cause SHORT syndrome. Clin Genet. 2014;85(3):228–9.
Buckley RH, Schiff SE, Sampson HA, Schiff RI, Markert ML, Knutsen AP, et al. Development of immunity in human severe primary T cell deficiency following haploidentical bone marrow stem cell transplantation. J Immunol. 1986;136:2398–407.
Sugden B, Mark W. Clonal transformation of adult human leukocytes by Epstein-Barr virus. J Virol. 1977;23(3):503–8.
Gorentla BK, Wan CK, Zhong XP. Negative regulation of mTOR activation by diacylglycerol kinases. Blood. 2011;117(15):4022–31.
Seyama K, Nonoyama S, Gangsaas I, Hollenbaugh D, Pabst HF, Aruffo A, et al. Mutations of the CD40 ligand gene and its effect on CD40 ligand expression in patients with X-linked hyper IgM syndrome. Blood. 1998;92(7):2421–34.
Elgizouli M, Lowe DM, Speckmann C, Schubert D, Hulsdunker J, Eskandarian Z, et al. Activating PI3Kdelta mutations in a cohort of 669 patients with primary immunodeficiency. Clin Exp Immunol. 2015;6.
Burke JE, Williams RL. Synergy in activating class I PI3Ks. Trends Biochem Sci. 2015;40(2):88–100.
Geering B, Cutillas PR, Vanhaesebroeck B. Regulation of class IA PI3Ks: is there a role for monomeric PI3K subunits? Biochem Soc Trans. 2007;35(Pt 2):199–203.
Chiu YH, Lee JY, Cantley LC. BRD7, a tumor suppressor, interacts with p85alpha and regulates PI3K activity. Mol Cell. 2014;54(1):193–202.
Cheng Z, Tseng Y, White MF. Insulin signaling meets mitochondria in metabolism. Trends Endocrinol Metab. 2010;21(10):589–98.
Winnay JN, Solheim MH, Dirice E, Sakaguchi M, Noh HL, Kang HJ, et al. PI3-kinase mutation linked to insulin and growth factor resistance in vivo. J Clin Invest. 2016. doi:10.1172/JCI84005.
Yang J, Zhang P, Krishna S, Wang J, Lin X, Huang H, et al. Unexpected positive control of NFkappaB and miR-155 by DGKalpha and zeta ensures effector and memory CD8+ T Cell differentiation. Oncotarget. 2016. doi:10.18632/oncotarget.8164.
Petrovski S, Wang Q, Heinzen EL, Allen AS, Goldstein DB. Genic intolerance to functional variation and the interpretation of personal genomes. PLoS Genet. 2013;9(8):e1003709.
Blake JA, Bult CJ, Eppig JT, Kadin JA, Richardson JE. The mouse genome database: integration of and access to knowledge about the laboratory mouse. Nucleic Acids Res. 2014;42(Database issue):D810–7.
Zhu X, Petrovski S, Xie P, Ruzzo EK, Lu YF, McSweeney KM, et al. Whole-exome sequencing in undiagnosed genetic diseases: interpreting 119 trios. Genet Med. 2015;17:774–81.
Acknowledgments
This study was supported by the following grants: Baxter Healthcare Grant BT13-21694 to R.H.B. and NIH Grant R01 AI101206 to X-P. Z. Partial funding for this study was provided by UCB Celltech, including salary support for D.B.G. M.A. and D.M. are employees of UCB. The other authors declare no conflict of interest. S.P. is a National Health and Medical Research Council (NHMRC) CJ Martin Fellow. We thank our patients and their parents for participating in this study. We also thank B Krueger, J Bridges, Q Wang, N Ren, S Gewalt, S Kisselev, Y-F Lu, K Cronin, and N Walley for excellent technical support. We would like to acknowledge the following individuals for the contributions of control samples: W. B. Gallentine, E.L. Heinzen, A.M. Husain, K. N. Linney, M. A. Mikati, R. A. Radtke, and S. R. Sinha; J.P. McEvoy, A. Need, J. Silver, and M. Silver; D. H. Murdock and The MURDOCK Study Community Registry and Biorepository; G. Cavalleri, N. Delanty, and C. Depondt; J. Burke, C. Hulette, and K. Welsh-Bohmer; J. Milner; J. Hoover-Fong, N. L. Sobreira and D. Valle; E. J. Holtzman; W. L. Lowe; P. Lugar; S. M. Palmer; Z. Farfel, D. Lancet, E. Elon Pras; A. Poduri; M. Hauser; D. Marchuk; D. Koltai Attix, O. Chiba-Falek; E. T. Cirulli, V. Dixon and J. McEvoy; K. Schmader, S. McDonald, H. K. White, M. Yanamadala, and the Carol Woods and Croasdale Retirement Communities; R. Gbadegesin and M. Winn; D. Daskalakis; Q. Zhao; A. Holden and E. Behr; R. Brown; and S. Kerns and H. Oster. The collection of control samples was funded in part by Bryan ADRC NIA P30 AG028377, the Ellison Medical Foundation New Scholar award AG-NS-0441-08, an award from SAIC-Frederick, Inc. (M11-074), funding from Biogen Idec, NIMH awards RC2MH089915, R01MH097971, R01MH099216, and K01MH098126, the Epi4K Gene Discovery in Epilepsy study (NINDS U01-NS077303) and the Epilepsy Genome/Phenome Project (EPGP - NINDS U01-NS053998), and the Center for HIV/AIDS Vaccine Immunology (“CHAVI”) under a grant from the National Institute of Allergy and Infectious Diseases, National Institutes of Health (UO1AIO67854), NIAID (R01AI079088 and R01AI101206). Finally, the authors would also like to thank the Exome Aggregation Consortium and the groups that provided exome variant data for comparison. A full list of contributing groups can be found at http://exac.broadinstitute.org/about.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
ESM 1
Figure S1: Sanger sequencing traces of DNA sequence around the PIK3R1 donor splice site for each patient (1–4) and their parents (M = Mother, F = Father). Both forward (+) and reverse (−) directions are provided. Patients are heterozygous for the variant. The variant was not observed in the unaffected parents. Father of Patient 4 was unavailable. (JPEG 2560 kb)
Rights and permissions
About this article
Cite this article
Petrovski, S., Parrott, R.E., Roberts, J.L. et al. Dominant Splice Site Mutations in PIK3R1 Cause Hyper IgM Syndrome, Lymphadenopathy and Short Stature. J Clin Immunol 36, 462–471 (2016). https://doi.org/10.1007/s10875-016-0281-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10875-016-0281-6