Abstract
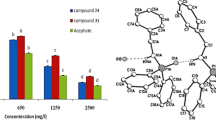
The development of new, more selective, environmental-friendly insecticide alternatives is in high demand for the control of Spodoptera frugiperda (S. frugiperda). The major objective of this work was to search for new potential S. frugiperda acetylcholinesterase (AChE) inhibitors. A ligand-based virtual screening was initially carried out considering six scaffolds derived from eugenol and the ZINC15, PubChem, and MolPort databases. Subsequently, molecular docking analysis of the selected compounds on the active site and a second region (determined by blind molecular docking) of the AChE of S. frugiperda was performed. Molecular dynamics and Molecular Mechanics Poisson–Boltzmann Surface Area analyses were also applied to improve the docking results. Finally, three new eugenol analogs were evaluated in vitro against S. frugiperda larvae. The virtual screening identified 1609 compounds from the chemical libraries. Control compounds were selected from the interaction fingerprint by molecular docking. Only three new eugenol analogs (1, 3, and 4) were stable at 50 ns by molecular dynamics. Compounds 1 and 4 had the best biological activity by diet (LC50 = 0.042 mg/mL) and by topical route (LC50 = 0.027 mg/mL), respectively. At least three new eugenol derivatives possessed good-to-excellent insecticidal activity against S. frugiperda.
Graphic abstract





source Version 2.4





Similar content being viewed by others
Data availability
Data are available from the authors upon reasonable request.
References
EPPO (2020) European and Mediterranean Plant Protection Organization. Spodoptera frugiperda. https://gd.eppo.int/taxon/LAPHFR. Accessed 15 Mar 2020
FAO (2020) Food and Agriculture Organization of the United Nations. FAOSTAT, Data, Production, Crops. http://www.fao.org/faostat/en/#compare. Accessed 08 Nov 2020
Blanco CA, Pellegaud JG, Nava-Camberos U, Lugo-Barrera D, Vega-Aquino P, Coello J et al (2014) Maize pests in Mexico and challenges for the adoption of integrated pest management programs. J Integr Pest Manag 5(4):E1–E9. https://doi.org/10.1603/IPM14006
IRAC (2020) Lepidoptera Insecticide Mode of Action Classification: A key to effective insecticide resistance management. https://irac-online.org/lepidoptera/. Accessed 03 Sept 2020
Sparks TC, Nauen R (2015) IRAC: mode of action classification and insecticide resistance management. Pest Biochem Physiol 121:122–128. https://doi.org/10.1016/j.pestbp.2014.11.014
Prasanna B, Huesing J, Eddy R, Peschke V (2018) Fall armyworm in Africa: a guide for integrated pest management, 1st edn. CIMMYT, Mexico, CDMX
Dvir H, Silman I, Harel M, Rosenberry TL, Sussman JL (2010) Acetylcholinesterase: from 3D structure to function. Chem Biol Interact 187(1–3):10–22. https://doi.org/10.1016/j.cbi.2010.01.042
Hernández-Carlos B, Gamboa-Angulo M (2019) Insecticidal and nematicidal contributions of Mexican Flora in the search for safer biopesticides. Molecules 24(5):897. https://doi.org/10.3390/molecules24050897
Cruz GS, Wanderley-Teixeira V, Oliveira JV, D’assunção CG, Cunha FM, Teixeira ÁA et al (2017) Effect of trans-anethole, limonene and your combination in nutritional components and their reflection on reproductive parameters and testicular apoptosis in Spodoptera frugiperda (Lepidoptera: Noctuidae). Chem Biol Interact 263:74–80. https://doi.org/10.1016/j.cbi.2016.12.013
Melani D, Himawan T, Afandhi A (2016) Bioactivity of sweet flag (Acorus calamus Linnaeus) essential oils against Spodoptera litura Fabricius (Lepidoptera: Noctuidae). J Trop Life Sci 6(2):86–90. https://doi.org/10.11594/jtls.06.02.04
Menichini F, Tundis R, Loizzo MR, Bonesi M, Marrelli M, Statti GA et al (2009) Acetylcholinesterase and butyrylcholinesterase inhibition of ethanolic extract and monoterpenes from Pimpinella anisoides V Brig. (Apiaceae). Fitoterapia 80(5):297–300. https://doi.org/10.1016/j.fitote.2009.03.008
Vargas-Méndez LY, Sanabria-Flórez PL, Saavedra-Reyes LM, Merchan-Arenas DR, Kouznetsov VV (2018) Bioactivity of semisynthetic eugenol derivatives against Spodoptera frugiperda (Lepidoptera: Noctuidae) larvae infesting maize in Colombia. Saudi J Biol Sci. https://doi.org/10.1016/j.sjbs.2018.09.010
Mukherjee PK, Kumar V, Mal M, Houghton PJ (2007) In vitro acetylcholinesterase inhibitory activity of the essential oil from Acorus calamus and its main constituents. Planta Med 73(03):283–285. https://doi.org/10.1055/s-2007-967114
O’Boyle NM, Banck M, James CA, Morley C, Vandermeersch T, Hutchison GR (2011) Open Babel: an open chemical toolbox. J Cheminform 3(1):33. https://doi.org/10.1186/1758-2946-3-33
Tice CM (2001) Selecting the right compounds for screening: does Lipinski’s Rule of 5 for pharmaceuticals apply to agrochemicals? Pest Manag Sci Former Pest Sci 57(1):3–16. https://doi.org/10.1002/1526-4998(200101)57:1%3c3::AID-PS269%3e3.0.CO;2-6
Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R et al (2018) SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res 46(W1):W296–W303. https://doi.org/10.1093/nar/gky427
Laskowski RA, MacArthur MW, Moss DS, Thornton JM (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr 26(2):283–291. https://doi.org/10.1107/S0021889892009944
Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC et al (2004) UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25(13):1605–1612. https://doi.org/10.1002/jcc.20084
Forli S, Huey R, Pique ME, Sanner MF, Goodsell DS, Olson AJ (2016) Computational protein–ligand docking and virtual drug screening with the AutoDock suite. Nat Protoc 11(5):905–919. https://doi.org/10.1038/nprot.2016.051
Trott O, Olson AJ (2010) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 31(2):455–461. https://doi.org/10.1002/jcc.21334
ChemAxon (2020) Marvin. https://chemaxon.com/products/marvin. Accessed 03 Feb 2020
Seeliger D, de Groot BL (2010) Ligand docking and binding site analysis with PyMOL and Autodock/Vina. J Comput Aided Mol Des 24(5):417–422. https://doi.org/10.1007/s10822-010-9352-6
Wójcikowski M, Zielenkiewicz P, Siedlecki P (2015) Open drug discovery toolkit (ODDT): a new open-source player in the drug discovery field. J Cheminform 7(1):1–6. https://doi.org/10.1186/s13321-015-0078-2
Chupakhin V, Marcou G, Gaspar H, Varnek A (2014) Simple ligand-receptor interaction descriptor (SILIRID) for alignment-free binding site comparison. Comput Struct Biotechnol J 10(16):33–37. https://doi.org/10.1016/j.csbj.2014.05.004
Baptista LPR, Sinatti VV, Da Silva JH, Dardenne LE, Guimarães AC (2019) Computational evaluation of natural compounds as potential inhibitors of human PEPCK-M: an alternative for lung cancer therapy. Adv Appl Bioinform Chem 12:15. https://doi.org/10.2147/AABC.S197119
Salentin S, Schreiber S, Haupt VJ, Adasme MF, Schroeder M (2015) PLIP: fully automated protein–ligand interaction profiler. Nucleic Acids Res 43(W1):W443–W447. https://doi.org/10.1093/nar/gkv315
Abraham MJ, Murtola T, Schulz R, Páll S, Smith JC, Hess B et al (2015) GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1:19–25. https://doi.org/10.1016/j.softx.2015.06.001
Dodda LS, Cabeza de Vaca I, Tirado-Rives J, Jorgensen WL (2017) LigParGen web server: an automatic OPLS-AA parameter generator for organic ligands. Nucleic Acids Res 45(W1):W331–W336. https://doi.org/10.1093/nar/gkx312
Polishchuk P, Kutlushina A, Bashirova D, Mokshyna O, Madzhidov T (2019) Virtual screening using pharmacophore models retrieved from molecular dynamic simulations. Int J Mol Sci 20(23):5834. https://doi.org/10.3390/ijms20235834
Lemkul J (2018) From proteins to perturbed Hamiltonians: a suite of tutorials for the GROMACS-2018 molecular simulation package [article v1. 0]. Living J Comput Mole Sci 1(1):5068. https://doi.org/10.33011/livecoms.1.1.5068
Kumari R, Kumar R, Lynn A (2014) g_mmpbsa—A GROMACS tool for high-throughput MM-PBSA calculations. J Chem Inf Model 54(7):1951–62. https://doi.org/10.1021/ci500020m
Liu J-Y, Chen X-E, Zhang Y-L (2015) Insights into the key interactions between human protein phosphatase 5 and cantharidin using molecular dynamics and site-directed mutagenesis bioassays. Sci Rep 5(1):1–11. https://doi.org/10.1038/srep12359
Rosado-Solano DN, Sanabria-Florez PL, Barón-Rodríguez MA, Luna-Parada LK, Puerto Galvis CE, Zorro-González AF et al (2019) Synthesis, biological evaluation and in silico computational studies of 7-Chloro-4-(1H–1, 2, 3-triazol-1-yl) quinoline derivatives. Search for new controlling agents against Spodoptera frugiperda (Lepidoptera: Noctuidae) larvae. J Agric Food Chem. https://doi.org/10.1021/acs.jafc.9b01067
Benkert P, Biasini M, Schwede T (2011) Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics 27(3):343–350. https://doi.org/10.1093/bioinformatics/btq662
Velankar S, Alhroub Y, Best C, Caboche S, Conroy MJ, Dana JM, et al (2012) PDBe: Protein Data Bank in Europe. Nucleic Acids Res. https://doi.org/10.1093/nar/gkr998
Haddad Y, Adam V, Heger Z (2020) Ten quick tips for homology modeling of high-resolution protein 3D structures. PLoS Comput Biol 16(4):e1007449. https://doi.org/10.1371/journal.pcbi.1007449
Xiang Z (2006) Advances in homology protein structure modeling. Current Protein Peptide Sci 7(3):217–227. https://doi.org/10.2174/138920306777452312
Fortney K, Griesman J, Kotlyar M, Pastrello C, Angeli M, Sound-Tsao M et al (2015) Prioritizing therapeutics for lung cancer: an integrative meta-analysis of cancer gene signatures and chemogenomic data. PLoS Comput Biol 11(3):e1004068. https://doi.org/10.1371/journal.pcbi.1004068
Nguyen NT, Nguyen TH, Pham TNH, Huy NT, Bay MV, Pham MQ et al (2019) Autodock vina adopts more accurate binding poses but autodock4 forms better binding affinity. J Chem Inf Model 60(1):204–211. https://doi.org/10.1021/acs.jcim.9b00778
Bhowmik D, Jagadeesan R, Rai P, Nandi R, Gugan K, Kumar D (2020) Evaluation of potential drugs against leishmaniasis targeting catalytic subunit of Leishmania donovani nuclear DNA primase using ligand based virtual screening, docking and molecular dynamics approaches. J Biomole Struct Dyn. https://doi.org/10.1080/07391102.2020.1739557
Kumari M, Subbarao N (2020) Virtual screening to identify novel potential inhibitors for Glutamine synthetase of Mycobacterium tuberculosis. J Biomole Struct Dyn 38(17):5062–5080. https://doi.org/10.1080/07391102.2019.1695670
Liao KH, Chen K-B, Lee W-Y, Sun M-F, Lee C-C, Chen CY-C (2014) Ligand-based and structure-based investigation for Alzheimer’s disease from traditional Chinese medicine. Evidence-Based Complement Altern Med. https://doi.org/10.1155/2014/364819
Rampogu S, Son M, Park C, Kim H-H, Suh J-K, Lee KW (2017) Sulfonanilide derivatives in identifying novel aromatase inhibitors by applying docking, virtual screening, and MD simulations studies. Biomed Res Int. https://doi.org/10.1155/2017/2105610
Zhang X, Yan J, Wang H, Wang Y, Wang J, Zhao D (2020) Molecular docking, 3D-QSAR, and molecular dynamics simulations of thieno [3, 2-b] pyrrole derivatives against anticancer targets of KDM1A/LSD1. J Biomole Struct Dyn. https://doi.org/10.1080/07391102.2020.1726819
Torres P, Avila JG, de Vivar AR, Garcia AM, Marin JC, Aranda E et al (2003) Antioxidant and insect growth regulatory activities of stilbenes and extracts from Yucca periculosa. Phytochemistry 64(2):463–73. https://doi.org/10.1016/S0031-9422(03)00348-0
Funding
The authors have no relevant financial or non-financial interests to disclose.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All authors certify that they have no affiliations with or involvement in any organization or entity with any financial interest or non-financial interest in the subject matter or materials discussed in this manuscript.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Méndez-Álvarez, D., Herrera-Mayorga, V., Juárez-Saldivar, A. et al. Ligand-based virtual screening, molecular docking, and molecular dynamics of eugenol analogs as potential acetylcholinesterase inhibitors with biological activity against Spodoptera frugiperda. Mol Divers 26, 2025–2037 (2022). https://doi.org/10.1007/s11030-021-10312-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11030-021-10312-5