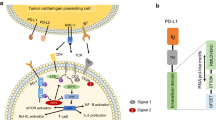
Abstract
Immunotherapy is widely used to treat various cancers, and the drugs used are called immune checkpoint (ICP) inhibitors. Overexpression of immune cell checkpoints is reported for other human diseases such as acute infections (malaria), chronic viral infection (HIV, hepatitis B virus, TB infections), allergy, asthma, neurodegeneration, and autoimmune diseases. Some mAbs (monoclonal antibodies) are available against ICPs, but they have side effects. Small molecule seems to be safer in comparison with mAbs. Three independent small-molecule inhibitor libraries consisting of 9466 compounds were screened against seven immune cell checkpoints by applying high-throughput virtual screening approach. A total of 13 ICP inhibitors were finalized based on docking, MM-GBSA scores, and ADME properties. Six compounds were selected for MD simulation, and then, rutin hydrate (targeting all seven immune cell checkpoints), amikacin hydrate (targeting six), and 6-hydroxyluteolin (targeting three) were found to be the best immune cell checkpoint inhibitors. These three potential inhibitors have shown the potential to activate human immune cells and thus may control the spread of human lifestyle or infectious diseases. Proposed inhibitors warrant the in vitro and in vivo validation to develop it as an immunotherapeutic.
Graphical abstract












Similar content being viewed by others
References
Pardoll DM (2012) The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer 12(4):252–264
Illingworth J, Butler NS, Roetynck S, Mwacharo J, Pierce SK, Bejon P, Crompton PD, Marsh K, Ndungu FM (2013) Chronic exposure to plasmodium falciparum is associated with phenotypic evidence of B and T cell exhaustion. J Immunol 190(3):1038–1047
Gonçalves-Lopes RM, Lima NF, Carvalho KI, Scopel KK, Kallás EG, Ferreira MU (2016) Surface expression of inhibitory (CTLA-4) and stimulatory (OX40) receptors by CD4+ regulatory T cell subsets circulating in human malaria. Microbes Infect 18(10):639–648
Trautmann L, Janbazian L, Chomont N, Said EA, Gimmig S, Bessette B, Boulassel MR, Delwart E, Sepulveda H, Balderas RS, Routy JP (2006) Upregulation of PD-1 expression on HIV-specific CD8+ T cells leads to reversible immune dysfunction. Nat Med 12(10):1198–1202
Chew GM, Fujita T, Webb GM, Burwitz BJ, Wu HL, Reed JS, Hammond KB, Clayton KL, Ishii N, Abdel-Mohsen M, Liegler T (2016) TIGIT marks exhausted T cells, correlates with disease progression, and serves as a target for immune restoration in HIV and SIV infection. PLoS Pathog 12(1):e1005349
Pollock KM, Montamat-Sicotte DJ, Grass L, Cooke GS, Kapembwa MS, Kon OM, Sampson RD, Taylor GP, Lalvani A (2016) PD-1 expression and cytokine secretion profiles of mycobacterium tuberculosis-specific CD4+ T-cell subsets; potential correlates of containment in HIV-TB co-infection. PLoS ONE 11(1):e0146905
Wykes MN, Lewin SR (2018) Immune checkpoint blockade in infectious diseases. Nat Rev Immunol 18(2):91–104
Lipson EJ, Drake CG (2011) Ipilimumab: an anti-CTLA-4 antibody for metastatic melanoma. Clin Cancer Res 17(22):6958–6962
Vial T, Choquet-Kastylevsky G, Descotes J (2002) Adverse effects of immunotherapeutics involving the immune system. Toxicology 174(1):3–11
The_American_Cancer_Society_medical_and_editorial_content_team, Monoclonal antibodies and their side effects. American Cancer Society, 2019
Sansom DM (2000) CD28, CTLA-4 and their ligands: Who does what and to whom? Immunology 101(2):169
Olsson C, Michaëlsson E, Parra E, Pettersson U, Lando PA, Dohlsten M (1998) Biased dependency of CD80 versus CD86 in the induction of transcription factors regulating the human IL-2 promoter. Int Immunol 10(4):499–506
van der Merwe PA, Bodian DL, Daenke S, Linsley P, Davis SJ (1997) CD80 (B7–1) binds both CD28 and CTLA-4 with a low affinity and very fast kinetics. J Exp Med 185(3):393–404
Linsley PS, Greene JL, Brady W, Bajorath J, Ledbetter JA, Peach R (1994) Human B7–1 (CD80) and B7–2 (CD86) bind with similar avidities but distinct kinetics to CD28 and CTLA-4 receptors. Immunity 1(9):793–801
Keir ME, Butte MJ, Freeman GJ, Sharpe AH (2008) PD-1 and its ligands in tolerance and immunity. Annu Rev Immunol 26:677–704
Liang SC, Latchman YE, Buhlmann JE, Tomczak MF, Horwitz BH, Freeman GJ, Sharpe AH (2003) Regulation of PD-1, PD-L1, and PD-L2 expression during normal and autoimmune responses. Eur J Immunol 33(10):2706–2716
Almahmoud S, Zhong HA (2019) Molecular modeling studies on the binding mode of the PD-1/PD-L1 complex inhibitors. Int J Mol Sci 20(18):4654
Nishimura H, Nose M, Hiai H, Minato N, Honjo T (1999) Development of lupus-like autoimmune diseases by disruption of the PD-1 gene encoding an ITIM motif-carrying immunoreceptor. Immunity 11(2):141–151
Raedler LA (2015) Keytruda (pembrolizumab): first PD-1 inhibitor approved for previously treated unresectable or metastatic melanoma. Am Health Drug Benefits 8(1):96
Monney L, Sabatos CA, Gaglia JL, Ryu A, Waldner H, Chernova T, Manning S, Greenfield EA, Coyle AJ, Sobel RA, Freeman GJ (2002) Th1-specific cell surface protein Tim-3 regulates macrophage activation and severity of an autoimmune disease. Nature 415(6871):536–541
Wolf Y, Anderson AC, Kuchroo VK (2020) TIM3 comes of age as an inhibitory receptor. Nat Rev Immunol 20(3):173–185
Murtaza A, Laken H, Correia JDS, McNeeley P, Altobell L, Zhang J, Vancutsem P, Wilcoxen K, Jenkins D (2016) Discovery of TSR-022, a novel, potent anti-human TIM-3 therapeutic antibody. Eur J Cancer 1(69):S102
Klein C, Schaefer W, Regula JT, Dumontet C, Brinkmann U, Bacac M, Umaña P (2019) Engineering therapeutic bispecific antibodies using CrossMab technology. Methods 154:21–31
Yu X, Harden K, Gonzalez LC, Francesco M, Chiang E, Irving B, Tom I, Ivelja S, Refino CJ, Clark H, Eaton D (2009) The surface protein TIGIT suppresses T cell activation by promoting the generation of mature immunoregulatory dendritic cells. Nat Immunol 10(1):48–57
Dolgin E (2020) Antibody engineers seek optimal drug targeting TIGIT checkpoint. Nat Biotechnol 38(9):1007–1010
Gonzalez LC, Loyet KM, Calemine-Fenaux J, Chauhan V, Wranik B, Ouyang W, Eaton DL (2005) A coreceptor interaction between the CD28 and TNF receptor family members B and T lymphocyte attenuator and herpesvirus entry mediator. Proc Natl Acad Sci 102(4):1116–1121
Rajalingam R (2012) Overview of the killer cell immunoglobulin-like receptor system. Immunogenetics, 391–414
Campbell KS, Purdy AK (2011) Structure/function of human killer cell immunoglobulin-like receptors: lessons from polymorphisms, evolution, crystal structures and mutations. Immunology 132(3):315–325
Núñez F, Taura J, Camacho J, López-Cano M, Fernández-Dueñas V, Castro N, Castro J, Ciruela F (2018) PBF509, an adenosine A2A receptor antagonist with efficacy in rodent models of movement disorders. Front Pharmacol, 1200
Furukawa, F. (2018) The Nobel prize in physiology or medicine 2018 was awarded to cancer therapy by inhibition of negative immune regulation. Trends Immunother 2(1)
Zang X (2018) 2018 Nobel prize in medicine awarded to cancer immunotherapy: Immune checkpoint blockade–a personal account. Genes Dis 5(4):302
Larkin J, Chiarion-Sileni V, Gonzalez R, Grob JJ, Cowey CL, Lao CD, Schadendorf D, Dummer R, Smylie M, Rutkowski P, Ferrucci PF (2015) Combined nivolumab and ipilimumab or monotherapy in untreated melanoma. N Engl J Med 373(1):23–34
Sznol M, Kluger HM, Callahan MK, Postow MA, Gordon RA, Segal NH, Rizvi NA, Lesokhin AM, Atkins MB, Kirkwood JM, Burke MM (2014) Survival, response duration, and activity by BRAF mutation (MT) status of nivolumab (NIVO, anti-PD-1, BMS-936558, ONO-4538) and ipilimumab (IPI) concurrent therapy in advanced melanoma (MEL)
Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE (2000) The protein data bank. Nucleic Acids Res 28(1):235–242
Gandhi AK, Kim WM, Sun ZYJ, Huang YH, Bonsor DA, Sundberg EJ, Kondo Y, Wagner G, Kuchroo VK, Petsko G, Blumberg RS (2018) High resolution X-ray and NMR structural study of human T-cell immunoglobulin and mucin domain containing protein-3. Sci Rep 8(1):1–13
Stengel KF, Harden-Bowles K, Yu X, Rouge L, Yin J, Comps-Agrar L, Wiesmann C, Bazan JF, Eaton DL, Grogan JL (2012) Structure of TIGIT immunoreceptor bound to poliovirus receptor reveals a cell–cell adhesion and signaling mechanism that requires cis-trans receptor clustering. Proc Natl Acad Sci 109(14):5399–5404
Moradi S, Berry R, Pymm P, Hitchen C, Beckham SA, Wilce MC, Walpole NG, Clements CS, Reid HH, Perugini MA, Brooks AG (2015) The structure of the atypical killer cell immunoglobulin-like receptor, KIR2DL4. J Biol Chem 290(16):10460–10471
Compaan DM, Gonzalez LC, Tom I, Loyet KM, Eaton D, Hymowitz SG (2005) Attenuating lymphocyte activity: the crystal structure of the BTLA-HVEM complex. J Biol Chem 280(47):39553–39561
Ramagopal UA, Liu W, Garrett-Thomson SC, Bonanno JB, Yan Q, Srinivasan M, Wong SC, Bell A, Mankikar S, Rangan VS, Deshpande S (2017) Structural basis for cancer immunotherapy by the first-in-class checkpoint inhibitor ipilimumab. Proc Natl Acad Sci 114(21):E4223–E4232
Lin DYW, Tanaka Y, Iwasaki M, Gittis AG, Su HP, Mikami B, Okazaki T, Honjo T, Minato N, Garboczi DN (2008) The PD-1/PD-L1 complex resembles the antigen-binding Fv domains of antibodies and T cell receptors. Proc Natl Acad Sci 105(8):3011–3016
Doré AS, Robertson N, Errey JC, Ng I, Hollenstein K, Tehan B, Hurrell E, Bennett K, Congreve M, Magnani F, Tate CG (2011) Structure of the adenosine A2A receptor in complex with ZM241385 and the xanthines XAC and caffeine. Structure 19(9):1283–1293
Madhavi Sastry G, Adzhigirey M, Day T, Annabhimoju R, Sherman W (2013) Protein and ligand preparation: parameters, protocols, and influence on virtual screening enrichments. J Comput Aided Mol Des 27(3):221–234
Zeng X, Zhang P, He W, Qin C, Chen S, Tao L, Wang Y, Tan Y, Gao D, Wang B, Chen Z (2018) NPASS: natural product activity and species source database for natural product research, discovery and tool development. Nucleic Acids Res 46(D1):D1217–D1222
Selleckchem, FDA-approved drug library. https://www.selleckchem.com/screening/fda-approved-drug-library.html
Selleckchem, FDA-approved & passed phase I drug library. https://www.selleckchem.com/screening/fda-approved-passed-phase-i-drug-library.html
Shelley JC, Cholleti A, Frye LL, Greenwood JR, Timlin MR, Uchimaya M (2007) Epik: a software program for pK a prediction and protonation state generation for drug-like molecules. J Comput Aided Mol Des 21(12):681–691
Halgren TA (2009) Identifying and characterizing binding sites and assessing druggability. J Chem Inf Model 49(2):377–389
Halgren T (2007) New method for fast and accurate binding-site identification and analysis. Chem Biol Drug Des 69(2):146–148
Glide EP (2006) Docking and scoring incorporating a model of hydrophobic enclosure for protein−ligand complexes. J Med Chem 4:6177–6196
Friesner RA, Murphy RB, Repasky MP, Frye LL, Greenwood JR, Halgren TA, Sanschagrin PC, Mainz DT (2006) Extra precision glide: docking and scoring incorporating a model of hydrophobic enclosure for protein−ligand complexes. J Med Chem 49(21):6177–6196
Jacobson MP, Pincus DL, Rapp CS, Day TJ, Honig B, Shaw DE, Friesner RA (2004) A hierarchical approach to all-atom protein loop prediction. Proteins Struct Funct Bioinform 55(2):351–367
Li J, Abel R, Zhu K, Cao Y, Zhao S, Friesner RA (2011) The VSGB 2.0 model: a next generation energy model for high resolution protein structure modeling. Proteins Struct Funct Bioinform 79(10):2794–2812
Fawcett T (2006) An introduction to ROC analysis. Pattern Recogn Lett 27(8):861–874
Naik B, Mattaparthi VSK, Gupta N, Ojha R, Das P, Singh S, Prajapati VK, Prusty D (2021) Chemical system biology approach to identify multi-targeting FDA inhibitors for treating COVID-19 and associated health complications. J Biomol Struct Dyn 1–25
WebGRO for macromolecular simulations (https://simlab.uams.edu/)
Abraham MJ, Murtola T, Schulz R, Páll S, Smith JC, Hess B, Lindahl E (2015) GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1:19–25
Schüttelkopf AW, Van Aalten DM (2004) PRODRG: a tool for high-throughput crystallography of protein–ligand complexes. Acta Crystallogr D Biol Crystallogr 60(8):1355–1363
Oostenbrink C, Villa A, Mark AE, Van Gunsteren WF (2004) A biomolecular force field based on the free enthalpy of hydration and solvation: the GROMOS force-field parameter sets 53A5 and 53A6. J Comput Chem 25(13):1656–1676
Ghorbani A (2017) Mechanisms of antidiabetic effects of flavonoid rutin. Biomed Pharmacother 96:305–312
Habtemariam S (2016) Rutin as a natural therapy for Alzheimer’s disease: insights into its mechanisms of action. Curr Med Chem 23(9):860–873
Rosenberg CR, Fang X, Allison KR (2020) Potentiating aminoglycoside antibiotics to reduce their toxic side effects. PLoS ONE 15(9):e0237948
Sanders WE Jr, Hartwig C, Schnieder N, Cacciatore R, Valdez H (1982) Activity of amikacin against mycobacteria in vitro and in murine tuberculosis. Tubercle 63(3):201–208
Clinical Trials.gov, Inulin for infections in the intensive care unit.
Berry ED, Wells JE (2010) Escherichia coli O157: H7: recent advances in research on occurrence, transmission, and control in cattle and the production environment. Adv Food Nutr Res 60:67–117
Scholar E (2007) Neomycin. in xpharm: The Comprehensive Pharmacology Reference. https://doi.org/10.1016/B978-008055232-3.62258-5
Sykes JE, Papich MG (2013) Antiprotozoal drugs. Canine Feline Infect Dis 171:97–104
Lipinski CA (2004) Lead-and drug-like compounds: the rule-of-five revolution. Drug Discov Today Technol 1(4):337–341
Acknowledgements
SS is thankful to Central University of Rajasthan for providing fellowship. VKP is thankful to the Central University of Rajasthan for providing laboratory facility.
Author information
Authors and Affiliations
Contributions
Protocol was designed by SS and VKP. Methodology was performed by SS, KK, MP, and VKP. The manuscript was written by SS, KK, AS, AM, and VKP.
Corresponding author
Ethics declarations
Conflict of interest
Authors have declared no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Singh, S., Kumar, K., Panda, M. et al. High-throughput virtual screening of small-molecule inhibitors targeting immune cell checkpoints to discover new immunotherapeutics for human diseases. Mol Divers 27, 729–751 (2023). https://doi.org/10.1007/s11030-022-10452-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11030-022-10452-2