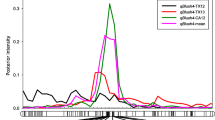
Abstract
Large fruit size is a critical trait for any new sweet cherry (Prunus avium L.) cultivar, as it is directly related to grower profitability. Therefore, determining the genetic control of fruit size in relevant breeding germplasm is a high priority. The objectives of this study were (1) to determine the number and positions of quantitative trait loci (QTL) for sweet cherry fruit size utilizing data simultaneously from multiple families and their pedigreed ancestors, and (2) to estimate fruit size QTL genotype probabilities and genomic breeding values for the plant materials. The sweet cherry material used was a five-generation pedigree consisting of 23 founders and parents and 424 progeny individuals from four full-sib families, which were phenotyped for fruit size and genotyped with 78 RosCOS single nucleotide polymorphism and 86 simple sequence repeat markers. These data were analyzed by a Bayesian approach implemented in FlexQTL™ software. Six QTL were identified: three on linkage group (G) 2 with one each on groups 1, 3, and 6. Of these QTL, the second G2 QTL and the G6 QTL were previously discovered while other QTL were novel. The predicted QTL genotypes show that some QTL were segregating in all families while other QTL were segregating in a subset of the families. The progeny varied for breeding value, with some progeny having higher breeding values than their parents. The results illustrate the use of multiple pedigree-linked families for integrated QTL mapping in an outbred crop to discover novel QTL and predict QTL genotypes and breeding values.



Similar content being viewed by others
References
Bink MCAM, Uimari P, Sillanpaa MJ, Janss LLG, Jansen RC (2002) Multiple QTL mapping in related plant populations via a pedigree-analysis approach. Theor Appl Genet 104:751–762
Bink MCAM, Boer MP, ter Braak CJF, Jansen J, Voorrips RE, van de Weg WE (2008) Bayesian analysis of complex traits in pedigreed plant populations. Euphytica 161:85–96
Bink MCAM, Radu Totir L, ter Braak CJF, Winkler CR, Boer MP, Smith OS (2012) QTL linkage analysis of connected populations using ancestral marker and pedigree information. Theor Appl Genet 124:1097–1113
Cabrera A, Kozik A, Howad W, Arus P, Iezzoni AF, van der Knaap E (2009) Development and bin mapping of a Rosaceae Conserved Ortholog Set (COS) of markers. BMC Genomics 10:562
Cabrera A, Rosyara UR, van der Knaap E, De Franceschi P, Sebolt A, Sooriyapathirana SS, Dirlewanger E, Quero-Garcia J, Schuster M, Iezzoni A (2012) Rosaceae COS SNP marker polymorphism in sweet cherry breeding germplasm and construction of a SNP-based map. Tree Genet Genomes 8:237–247
Clarke JB, Sargent DJ, Bošković RI, Belaj A, Tobutt KR (2009) A cherry map from the inter-specific cross Prunus avium ‘Napoleon’ × P. nipponica based on microsatellite gene-specific and isoenzyme markers. Tree Genet Genomes 5:41–51
De Franceschi P, Stegmeir T, Cabrera A, van der Knaap E, Rosyara UR, Sebolt AM, Dondini L, Dirlewanger E, Quero-Garcia J, Campoy JA, Iezzoni AF (2013) Cell number regulator genes in Prunus provide candidate genes for the control of fruit size in sweet and sour cherry. Mol Breed. doi:10.1007/s11032-013-9872-6
Dirlewanger E, Moing A, Rothan C, Svanella L, Pronier V, Guye A, Plomion C, Monet R (1999) Mapping QTLs controlling fruit quality in peach (Prunus persica (L.) Batsch). Theor Appl Genet 98:18–31
Dirlewanger E, Graziano E, Joobeur T, Garriga-Caldere F, Cosson P, Howad W et al (2004) Comparative mapping and marker-assisted selection in Rosaceae fruit crops. Proc Natl Acad Sci USA 101:9891–9896
Dirlewanger E, Quero-García J, Le Dantec L, Lambert P, Ruiz D, Dondini L, Illa E, Quilot-Turion B, Audergon JM, Tartarini S, Letourmy P, Arús P (2012) Comparison of the genetic determinism of two key phenologyical traits, flowering and maturity dates, in three Prunus species: peach, apricot and sweet cherry. Heredity 109:280–292
Eduardo I, Pacheco I, Chietera G, Bassi D, Pozzi C, Vecchietti A, Rossini L (2011) QTL analysis of fruit quality traits in two peach intraspecific populations and importance of maturity date pleiotropic effect. Tree Genet Genomes 7:323–335
Fogle HW (1961) Inheritance of some fruit and tree characteristics in sweet cherry crosses. Proc Am Soc Hort Sci 78:76–85
Gelman A, Rubin DB (1992) Inference from iterative simulation using multiple sequences. Stat Sci 7(4):472–475
Hurme P, Sillanpää MJ, Arjas E, Repo T, Savolainen O (2000) Genetic basis of climatic adaptation in Scots pine by Bayesian quantitative trait locus analysis. Genetics 156:1309–1322
Jannink J, Bink MCAM, Jansen RC (2001) Using complex plant pedigrees to map valuable genes. Trends Plant Sci 6:337–342
Jansen RC, Jannink J, Beavis WD (2003) Mapping quantitative trait loci in plant breeding populations: use of parental haplotype sharing. Crop Sci 43:829–834
Jansen J, Bink MCAM, Boer MP, Van de Weg WE (2009) Searching for interactions in related populations of an outbreeding species. Euphytica 166:131–144
Kass RE, Raftery AE (1995) Bayes factors. J Am Stat Assoc 90(430):773–795
Klagges C, Campoy JA, Quero-García J, Guzman A, Mansur L, Gratacós E, Silva H, Rosyara UR, Iezzoni A, Meisel L, Dirlewanger E (2013) Construction and comparative analyses of highly dense linkage maps of two sweet cherry intra-specific progenies of commercial cultivars. PLoS ONE 8(1):e54743
Lamb RC (1953) Notes on the inheritance of some characters in the sweet cherry Prunus avium. Proc Am Soc Hort Sci 61:293–298
Maliepaard C, Sillanpää MJ, Van Ooijen J, Jansen RC, Arjas E (2001) Bayesian versus frequentist analysis of multiple quantitative trait loci with an application to an outbred apple cross. Theor Appl Genet 103:1243–1253
Mathews P (1973) Some recent advances in sweet cherry genetics and breeding. In: Proceedings of EUCARPIA fruit section symposium 5 topic fruit breeding, Canterbury, pp 84–107
Olmstead JW, Sebolt AM, Cabrera A, Sooriyapathirana SS, Hammar S, Iriarte G, Wang D, Chen CY, van der Knaap E, Iezzoni AF (2008) Construction of an intra-specific sweet cherry (Prunus avium L.) genetic linkage map and synteny analysis with the Prunus reference map. Tree Genet Genomes 4:897–910
Plummer M, Best NG, Cowles K, Vines K (2006) CODA: convergence diagnosis and output analysis for MCMC. R News 6:7–11
Quero-Garcia J, Campoy JA, Joly J, Tauzin Y, Rosyara U, Iezzoni A, Dirlewanger E (2012) QTL detection for fruit weight, fruit firmness and fruit cracking tolerance in sweet cherry, Plant & Animal Genomes XX Conference, January 14–18, 2012. San Diego, CA
Quilot B, Wu BH, Kervella J, Genard M, Goulongne M, Moreau K (2004) QTL analysis of quality traits in an advanced backcross between Prunus persica cultivars and the wild relative species P. davidiana. Theor Appl Genet 109:884–897
Rosyara UR, Gonzalez-Hernandez JL, Glover KD, Gedye KR, Stein JM (2009) Family-based mapping of quantitative trait loci in plant breeding populations with resistance to Fusarium head blight in wheat as an illustration. Theor Appl Genet 118:1617–1631
Shoemaker JS, Painter IS, Weir BS (1999) Bayesian statistics in genetics. A guide for the uninitiated. Trends Genet 15:354–358
Sillanpää MJ, Arjas E (1998) Bayesian mapping of multiple quantitative trait loci from incomplete inbred line cross data. Genetics 148:1373–1388
Sillanpää MJ, Arjas E (1999) Bayesian mapping of multiple quantitative trait loci from incomplete outbred offspring data. Genetics 151:1605–1619
Sorensen D, Gianola D (2002) Likelihood, Bayesian, and MCMC methods in quantitative genetics. Springer, New York
Stephens DA, Fisch RD (1998) Bayesian analysis of quantitative trait locus data using reversible jump Markov chain Monte Carlo. Biometrics 54:1334–1347
Stephens DA, Smith AFM (1993) Bayesian inference in multipoint gene mapping. Ann Hum Genet 57:65–82
R Development Core Team (2011) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. ISBN 3-900051-07-0, http://www.R-project.org
Thomas DC, Cortessis V (1992) A Gibbs sampling approach in linkage analysis. Hum Hered 42:63–76
Thompson EA (2008) Linkage analysis. In: Balding DJ, Bishop M, Cannings C (eds) Handbook of statistical genetics, 3rd edn. Wiley, Chichester, pp 1141–1167
van de Weg WE, Voorrips RE, Finkers HJ, Kodde LP, Meulenbroek EJ, Jansen J, Bink MCAM (2005) Pedigree genotyping: a new pedigree-based approach of QTL identification and allele mining by exploiting breeding material. Acta Hort 708:483–488
Voorrips RE (2002) MapChart: software for the graphical presentation of linkage maps and QTLs. J Hered 93:77–78
Voorrips RE, Bink MCAM, van de Weg WE (2012) Pedimap: software for the visualization of genetic and phenotypic data in pedigrees. J Hered 103:903–907
Whiting MD, Ophardt D, McFerson JR (2006) Chemical blossom thinners vary in their effect on sweet cherry fruit set, yield, fruit quality, and crop value. Hort Technol 16:66–70
Yi N, Xu S (2000) Bayesian mapping of quantitative trait loci under the identity-by-descent-based variance component model. Genetics 156:411–422
Zhang G, Sebolt AM, Sooriyapathirana SS, Wang D, Bink MCAM, Olmstead JW, Iezzoni A (2010) Fruit size QTL analysis of an F1 population derived from a cross between a domesticated sweet cherry cultivar and a wild forest sweet cherry. Tree Genet Genomes 6:25–36
Acknowledgments
This project was supported in part by two USDA grants, USDA-NRI NIFA 2008-02259 and USDA-SCRI Grant #2009 = 51181-05808 entitled ‘RosBREED: Enabling marker-assisted breeding in Rosaceae’.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Table S1
Populations, locations, and number of progeny individuals genotyped for 86 SSR and 78 RosCOS SNP markers. This plant material and genotyping is described in Cabrera et al (2012) (PPTX 99 kb)
Supplementary Table S2
Posterior probability for the fruit weight FW_G1, FW_G2a, FW_G2b, FW_G2c, FW_G3, and FW_G6 genotypes predicted by FlexQTL™, where Q is the positive QTL allele and q is negative QTL allele. Progeny from the bi-parental populations are identified by the combination of the first letter from both parents (PPTX 75 kb)
Supplementary Table S3
Estimated individual and genome-wide breeding values for the QTLs detected in this study. Progeny from the bi-parental populations are identified by the combination of the first letter from both parents (PPTX 67 kb)
Supplementary Figure S1
Full pedigree of the sweet cherry plant materials used in the study (PPTX 62 kb)
Supplementary Figure S2
Fruit weight (g) distributions of (1) all populations, (2) NY × EF (3) Regina × Lapins, (4) Namati × Krupnoplodnaya, and (5) Namati × Summit. n indicates number of individuals (PPTX 55 kb)
Supplementary Figure S3
Informative meioses at the first and second homologues across the eight sweet cherry linkage groups (PPTX 59 kb)
Supplementary Figure S4
Frequency of observed and expected double recombinants in the plant materials used in this study. The upper triangles indicate the first parent while the downward triangles indicate the second parent. The black and red colors are the expected and observed number of double recombinants, respectively. Stars result when the triangles from the first and second parent plot to the same position (DOCX 15 kb)
Supplementary Figure S5
QTL allele predictions (Q/q) and probabilities (QTL p) and breeding values (BV) for ‘Regina’ and ‘Lapins’ and two of their progeny individuals, RL0059 and RL0039, that have high and low breeding values, respectively. QTL genotypes predicted with a probability greater than 0.70 are in bold (XLSX 55 kb)
Supplementary Figure S6
Transmission for FW_G2b in progeny from the cross between ‘New York 54’ (New York) and ‘Emperor Francis’ (EF). The FW_G2b alleles are defined by the SSR marker BPPCT034. The predicted QTL genotypes are presented along with the fruit weight means for each of the four progeny genotypic classes. Fruit weigh means followed by the same letter are not significantly different (ANOVA, P > 0.05) (XLSX 167 kb)
Rights and permissions
About this article
Cite this article
Rosyara, U.R., Bink, M.C.A.M., van de Weg, E. et al. Fruit size QTL identification and the prediction of parental QTL genotypes and breeding values in multiple pedigreed populations of sweet cherry. Mol Breeding 32, 875–887 (2013). https://doi.org/10.1007/s11032-013-9916-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11032-013-9916-y