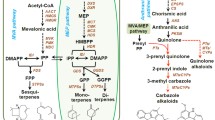
Abstract
Eugenol-O-methyltransferase (EOMT) catalyzes the conversion of eugenol to methyleugenol in one of the final steps of phenylpropanoid pathway. There are no comprehensive reports on comparative EOMT gene expression and developmental stage specific accumulation of phenylpropenes in Ocimum tenuiflorum. Seven chemotypes, rich in eugenol and methyleugenol, were selected by assessment of volatile metabolites through multivariate data analysis. Isoeugenol accumulated in higher levels during juvenile stage (36.86 ng g−1), but reduced sharply during preflowering (8.04 ng g−1), flowering (2.29 ng g−1) and postflowering stages (0.17 ng g−1), whereas methyleugenol content gradually increased from juvenile (12.25 ng g−1) up to preflowering (16.35 ng g−1) and then decreased at flowering (7.13 ng g−1) and post flowering (5.95 ng g−1) from fresh tissue. Extreme variations of free intracellular and alkali hydrolysable cell wall released phenylpropanoid compounds were observed at different developmental stages. Analyses of EOMT genomic and cDNA sequences revealed a 843 bp open reading frame and the presence of a 90 bp intron. The translated proteins had eight catalytic domains, the major two being dimerisation superfamily and methyltransferase_2 superfamily. A validated 3D structure of EOMT protein was also determined. The chemotype Ot7 had a reduced reading frame that lacked both dimerisation domains and one of the two protein-kinase-phosphorylation sites; this was also reflected in reduced accumulation of methyleugenol compared to other chemotypes. EOMT transcripts showed enhanced expression in juvenile stage that increased further during preflowering but decreased at flowering and further at postflowering. The expression patterns may possibly be compared and correlated to the amounts of eugenol/isoeugenol and methyleugenol in different developmental stages of all chemotypes.









Similar content being viewed by others
References
Simon JE, Morales MR, Phippen WB, Vieira RF, Hao Z (1999) Basil: a source of aroma compounds and a popular culinary and ornamental herb. In: Janick J (ed) Perspectives on new crops and new uses. ASHS Press, Alexandria, pp 499–505
WHO monographs on selected medicinal plants (2002) Folium Ocimi Sancti. 2:206–216
Agnieszka K, Kurowska A, Danuta K (2005) Composition of the essential oil of Ocimum sanctum L. grown in Poland during vegetation. J Essent Oil Res 17:217–219. doi:10.1080/10412905.2005.9698880
Lange BM, Wildung MR, Stauber EJ, Sanchez C, Pouchnik D, Croteau R (2000) Probing essential oil biosynthesis and secretion by functional evaluation of expressed sequence tags from mint glandular trichomes. Proc Natl Acad Sci USA 97:2934–2939. doi:10.1073/pnas.97.6.2934
Kothari SK, Bhattacharya AK, Ramesh S (2004) Essential oil yield and quality of methyl eugenol rich Ocimum tenuiflorum L.f. (syn. O. sanctum L.) grown in south India as influenced by method of harvest. J Chromatogr A 1054:67–72. doi:10.1016/j.chroma.2004.03.019
Vieira RF, Grayer RJ, Paton A, Simon JE (2001) Genetic diversity of Ocimum gratissimum L. based on volatile oil constituents, flavonoids and RAPD markers. Biochem Syst Ecol 29:287–304. doi:10.1016/S0305-1978(00)00062-4
Jirovetz L, Buchbauer G, Shafi MP, Kaniampady MM (2003) Chemotaxonomical analysis of the essential oils aroma compounds of four different Ocimum species from southern India. Eur Food Res Technol 217:120–124. doi:10.1007/s00217-003-0708-1
Viña A, Murillo E (2003) Essential oil composition from twelve varieties of Basil (Ocimum spp.) grown in Colombia. J Braz Chem Soc 14:744–749. doi:10.1590/S0103-50532003000500008
Dixon RA, Achnine L, Kota P, Liu C-J, Reddy MSS, Wang L (2002) The phenylpropanoid pathway and plant defence—a genomics perspective. Mol Plant Pathol 3:371–390. doi:10.1046/j.1364-3703.2002.00131.x
Hahlbrock K, Scheel D (1989) Physiology and molecular biology of phenylpropanoids metabolism. Annu Rev Plant Physiol Plant Mol Biol 40:347–369. doi:10.1146/annurev.arplant.40.1.347
Vogt T (2010) Phenylpropanoid biosynthesis. Mol Plant 3:2–20. doi:10.1093/mp/ssp106
Ibrahim RK, Bruneau A, Bantignies B (1998) Plant O-methyltransferases: molecular analysis, common signature and classification. Plant Mol Biol 36:1–10. doi:10.1023/A:1005939803300
Prakash P, Gupta N (2005) Therapeutic uses of Ocimum sanctum Linn (Tulsi) with a note on eugenol and its pharmacological actions: a short review. Indian J Physiol Pharmacol 49:125–131
Obeng-Ofori D, Reichmuth CH (1997) Bioactivity of eugenol, a major component of essential oil of Ocimum suave (wild) against four species of stored product Coleoptera. Int J Pest Manag 43:89–94. doi:10.1080/096708797229040
Wang J, Pichersky E (1999) Identification of specific residue involved in substrate discrimination in two plant O-methyltransferases. Arch Biochem Biophys 368:172–180. doi:10.1006/abbi1999.1304
Gang DR, Lavid N, Zubieta C, Chen F, Beuerle T, Lewinsohn E, Noel JP, Pichersky E (2002) Characterization of phenylpropene O-methyltransferases from sweet basil: facile changes of substrate specificity and convergent evolution within a plant O-methyltransferase family. Plant Cell 14:505–519. doi:10.1105/tpc.010327
Koeduka T, Fridman E, Gang DR, Vassa˜o DG, Jackson BL, Kish CM, Orlova I, Spassova SM, Lewis NG, Noel JP, Baiga TJ, Dudareva N, Pichersky E (2006) Eugenol and isoeugenol, characteristic aromatic constituents of spices, are biosynthesized via reduction of a coniferyl alcohol ester. Proc Natl Acad Sci USA 103:10128–10133. doi:10.1073/pnas.0603732103
Yamazaki M, Nakajima J, Yamanashi M, Sugiyama M, Makita Y, Springob K, Awazuhara M, Saito K (2003) Metabolomics and differential gene expression in anthocyanin chemo-varietal forms of Perilla frutescens. Phytochemistry 62:987–995. doi:10.1016/S0031-9422(02)00721-5
Carrari F, Baxter C, Usadel B, Urbanzyk-Wochniak E, Zanor MI, Nunes-Neri A, Nikiforova V, Centero D, Ratzka A, Pauly M, Sweetlove LJ, Fernie AR (2006) Integrated analysis of metabolite and transcript levels reveal the metabolite shifts that underlie tomato fruit development and highlight regulatory aspects of metabolic network behavior. Plant Physiol 142:1380–1396. doi:10.1104/pp.106.088534
Jumtee K, Bamba T, Okazawa A, Fukusaki E, Kobayashi A (2008) Integrated metabolite and gene expression profiling revealing phytochrome A regulation of polyamine biosynthesis of Arabidopsis thaliana. J Exp Bot 59:1187–1200. doi:10.1093/jxb/ern026
Roessner U, Bowne J (2009) What is metabolomics all about? Biotechniques 46:363–365. doi:10.2144/000113133
Iijima Y, Davidorich-Rikanati R, Fridman E, Gang DR, Bar E, Lewinsohn E, Pichersky E (2004) The biochemical and molecular basis for the divergent patterns in the biosynthesis of terpenes and phenylpropenes in the peltate glands of three cultivars of Basil. Plant Physiol 136:3724–3736. doi:10.1104/pp.104.051318
Tyssø V, Esbensen K, Martens H (1987) UNSCRAMBLER, an interactive program for multivariate calibration and prediction. Chemometr Intell Lab 2:239–249. doi:10.1016/0169-7439(87)80102-8
Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plantarum 15:473–497. doi:10.1111/j.1399-3054.1962.tb08052.x
Santiago R, Butron A, Arnason JT, Reid LM, Sauto XC, Malvar RA (2006) Putative role of pith cell wall phenylpropanoids in Sesamia nonagrioides (Lepidoptera: Noctuidae) resistance. J Agric Food Chem 54:2274–2279. doi:10.1021/jf0524271
Tan J, Bednarek P, Liu J, Schneider B, Svatoš A, Hahlbrock K (2004) Universally occurring phenylpropanoids and species specific indolic metabolites in infected and uninfected Arabidopsis thaliana roots and leaves. Phytochemistry 65:691–699. doi:10.1016/j.phytochem.2003.12.009
Haque I, Bandopadhyay R, Mukhopadhyay K (2008) An optimised protocol for fast genomic DNA isolation from high secondary metabolite and gum containing plants. Asian J Plant Sci 7:304–308. doi:10.3923/ajps.2008.304.308
Larkin MA, Blackshields G, Brown NP (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. doi:10.1093/bioinformatics/btm404
Felsenstein J (2002) Quantitative characters, phylogenies, and morphometrics, In: MacLeod N (ed) Morphology, shape and phylogenetics. Systematics Association 64:27–44. doi:10.1201/9780203165171.ch3
Jones DT, Taylor WR, Thornton JM (1992) The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci 8:275–282. doi:10.1093/bioinformatics/8.3.275
Saitou N, Nei M (1987) The neighbour–joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Naughton BT, Fratkin E, Batzoglou S, Brutlag DL (2006) A graph-based motif detection algorithm models complex nucleotide dependencies in transcription factor binding sites. Nucleic Acids Res 34:5730–5739. doi:10.1093/nar/gkl585
Šali A, Blundell T (1993) Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol 234:779–815. doi:10.1006/jmbi1993.1626
Wiederstein M, Sippl MJ (2007) ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res 35:407–410. doi:10.1093/nar/gkm290
Rein RS, Mauritz RP, Geyer M (2006) Universal ProbeLibrary: a new concept for streamlining gene expression analysis with qPCR. Nat Method Appl Notes. doi:10.1038/an1341
Horst I, Peterhänsel C (2007) Quantification of Zea mays mRNAs by real-time PCR using the Universal Probe Library. Biochemica 1:8–10
Paolacci AR, Tanzarella OA, Porceddu E, Ciaffi M (2009) Identification and validation of reference genes for quantitative RT-PCR normalization in wheat. BMC Mol Biol. doi:10.1186/1471-2199-10-11
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2 −ΔΔCT method. Methods 25:402–408. doi:10.1006/meth.2001.1262
Wang X (2011) Structure, function, and engineering of enzymes in isoflavonoid biosynthesis. Funct Integr Genomics 11:13–22. doi:10.1007/s10142-010-0197-9
Marchler-Bauer A, Anderson JB, Chitsaz F (2009) CDD: specific functional annotation with the conserved domain database. Nucleic Acids Res 37:205–210. doi:10.1093/nar/gkn845
Mellor H, Parker PJ (1998) The extended protein kinase C superfamily. Biochem J 332:281–292
Choi YH, Tapias EC, Kim HK, Lefeber AWM, Erkelens C, Verhoeven ThJJ, Brzin J, Zel J, Verpoorte R (2004) Metabolic discrimination of Catharanthus roseus leaves infected by phytoplasma using 1H-NMR spectroscopy and multivariate data analysis. Plant Physiol 135:2398–2410. doi:10.1104/pp.104.041012
Klejdus B, Vacek J, Lojková L, Benešová L, Kubáň V (2008) Ultrahigh-pressure liquid chromatography of isoflavones and phenolic acids on different stationary phases. J Chromatogr A 1195:52–59. doi:10.1016/j.chroma.2008.04.069
Quirantes-Piné R, Funes L, Micol V, Segura-Carretero A, Fernández-Gutiérrez A (2009) High-performance liquid chromatography with diode array detection coupled to electrospray time-of-flight and ion-trap tandem mass spectrometry to identify phenolic compounds from a lemon verbena extract. J Chromatogr A 1216:5391–5397. doi:10.1016/j.chroma.2009.05.038
Yoshida T, Majors RE (2006) High-speed analyses using rapid resolution liquid chromatography on 1.8-μm porous particles. J Sep Sci 29:2421–2432. doi:10.1002/jssc.200600267
Haque I, Kumar M, Mukhopadhyay K (2009) A rapid and simple UPLC-MS-MS based simultaneous determination of the medicinally important E- and Z-guggulsterone from oleo-gum resins of naturally occurring Commiphora wightii plants. Chromatographia 70:1613–1619. doi:10.1365/s10337-009-1347-x
Foyer CH, Noctor G, van Emden HF (2007) An evaluation of making specific secondary metabolites: does the yield penalty incurred by host plant resistance to insects result from competition for resources? Int J Pest Manag 53:175–182. doi:10.1080/09670870701469146
Acknowledgments
This work was supported by Departmental Grants of BIT Mesra. The authors are thankful to BTISNET SubDIC (BT/BI/04/065/04) for providing facilities for bioinformatics analyses and the Government of Jharkhand, Department of Agriculture (5/B.K.V/Misc/12/2001) for providing infrastructure development fund. Fellowships were provided to IKR by BIT-Mesra and IH by CSIR [9/554 (13) 2007-EMR-I].
Author information
Authors and Affiliations
Corresponding author
Additional information
Indu Kumari Renu, Inamul Haque contributed equally.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Renu, I.K., Haque, I., Kumar, M. et al. Characterization and functional analysis of eugenol O-methyltransferase gene reveal metabolite shifts, chemotype specific differential expression and developmental regulation in Ocimum tenuiflorum L.. Mol Biol Rep 41, 1857–1870 (2014). https://doi.org/10.1007/s11033-014-3035-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-014-3035-7