Abstract
Purpose
A circulating biomarker has potential to provide more accurate information for glioma progression post treatment, however no such biomarker is currently available. We aimed to discover a microRNA serum biomarker for longitudinal monitoring of glioma patients.
Methods
A prospectively collected cohort of 91 glioma patients and 17 healthy controls underwent pre and post-operative serum miRNA profiling using Nanostring®. Differentially expressed miRNAs were discovered using a machine learning random forest analysis. Candidate miRNAs were then assessed by droplet digital PCR in 11 patients with multiple follow up samples and compared to tumor volume based on magnetic resonance imaging.
Results
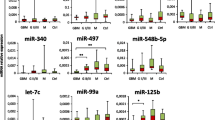
A 9-gene miRNA signature was identified that could distinguish between glioma and healthy controls with 99.8% accuracy. Two miRNAs miR-223 and miR-320e, best demonstrated dynamic changes that correlated closely with tumor volume in LGG and GBM respectively. Importantly, miRNA levels did not increase in two cases of pseudo-progression, indicating the potential utility of this test in guiding treatment decisions.
Conclusions
We identified a highly accurate 9-miRNA signature associated with glioma serum. Additionally, we observed dynamic changes in specific miRNAs correlating with tumor volume over long-term follow up. These results support a large prospective validation study of serum miRNA biomarkers in glioma.



Similar content being viewed by others
Data availability
All data generated or analysed during this study are included in this published article (and its supplementary information files).
Change history
07 October 2020
For the reference citation '[57]' in the second paragraph of the <Emphasis Type="Bold">Results</Emphasis> section of the original article there was no corresponding entry in the References section. It should have referred to the below mentioned article by Ebrahimkhani et al. (2018).
References
Kaye AH, Morokoff A (2014) The continuing evolution: biology and treatment of brain tumors. Neurosurgery 61(Suppl 1):100–104. https://doi.org/10.1227/NEU.0000000000000388
Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn U, Curschmann J, Janzer RC, Ludwin SK, Gorlia T, Allgeier A, Lacombe D, Cairncross JG, Eisenhauer E, Mirimanoff RO, European Organisation for R, Treatment of Cancer Brain T, Radiotherapy G, National Cancer Institute of Canada Clinical Trials G (2005) Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med 352:987–996. https://doi.org/10.1056/NEJMoa043330
Ellingson BM, Wen PY, Cloughesy TF (2017) Modified criteria for radiographic response assessment in glioblastoma clinical trials. Neurotherapeutics. https://doi.org/10.1007/s13311-016-0507-6
Sottoriva A, Spiteri I, Piccirillo SG, Touloumis A, Collins VP, Marioni JC, Curtis C, Watts C, Tavare S (2013) Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proc Natl Acad Sci USA 110:4009–4014. https://doi.org/10.1073/pnas.1219747110
Gourlay J, Morokoff AP, Luwor RB, Zhu HJ, Kaye AH, Stylli SS (2017) The emergent role of exosomes in glioma. J Clin Neurosci 35:13–23. https://doi.org/10.1016/j.jocn.2016.09.021
Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O'Briant KC, Allen A, Lin DW, Urban N, Drescher CW, Knudsen BS, Stirewalt DL, Gentleman R, Vessella RL, Nelson PS, Martin DB, Tewari M (2008) Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA 105:10513–10518. https://doi.org/10.1073/pnas.0804549105
Yang C, Wang C, Chen X, Chen S, Zhang Y, Zhi F, Wang J, Li L, Zhou X, Li N, Pan H, Zhang J, Zen K, Zhang CY, Zhang C (2013) Identification of seven serum microRNAs from a genome-wide serum microRNA expression profile as potential noninvasive biomarkers for malignant astrocytomas. Int J Cancer 132:116–127. https://doi.org/10.1002/ijc.27657
Zhi F, Shao N, Wang R, Deng D, Xue L, Wang Q, Zhang Y, Shi Y, Xia X, Wang S, Lan Q, Yang Y (2015) Identification of 9 serum microRNAs as potential noninvasive biomarkers of human astrocytoma. Neuro Oncol 17:383–391. https://doi.org/10.1093/neuonc/nou169
Roth P, Wischhusen J, Happold C, Chandran PA, Hofer S, Eisele G, Weller M, Keller A (2011) A specific miRNA signature in the peripheral blood of glioblastoma patients. J Neurochem 118:449–457. https://doi.org/10.1111/j.1471-4159.2011.07307.x
Dong L, Li Y, Han C, Wang X, She L, Zhang H (2014) miRNA microarray reveals specific expression in the peripheral blood of glioblastoma patients. Int J Oncol 45:746–756. https://doi.org/10.3892/ijo.2014.2459
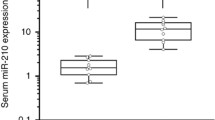
Lai NS, Wu DG, Fang XG, Lin YC, Chen SS, Li ZB, Xu SS (2015) Serum microRNA-210 as a potential noninvasive biomarker for the diagnosis and prognosis of glioma. Br J Cancer 112:1241–1246. https://doi.org/10.1038/bjc.2015.91
Regazzo G, Terrenato I, Spagnuolo M, Carosi M, Cognetti G, Cicchillitti L, Sperati F, Villani V, Carapella C, Piaggio G, Pelosi A, Rizzo MG (2016) A restricted signature of serum miRNAs distinguishes glioblastoma from lower grade gliomas. J Exp Clin Cancer Res 35:124. https://doi.org/10.1186/s13046-016-0393-0
Ma C, Nguyen HPT, Luwor RB, Stylli SS, Gogos A, Paradiso L, Kaye AH, Morokoff AP (2018) A comprehensive meta-analysis of circulation miRNAs in glioma as potential diagnostic biomarker. PLoS ONE 13:e0189452. https://doi.org/10.1371/journal.pone.0189452
Areeb Z, Stylli SS, Koldej R, Ritchie DS, Siegal T, Morokoff AP, Kaye AH, Luwor RB (2015) MicroRNA as potential biomarkers in glioblastoma. J Neurooncol 125:237–248. https://doi.org/10.1007/s11060-015-1912-0
Tabibkhooei A, Izadpanahi M, Arab A, Zare-Mirzaei A, Minaeian S, Rostami A, Mohsenian A (2020) Profiling of novel circulating microRNAs as a non-invasive biomarker in diagnosis and follow-up of high and low-grade gliomas. Clin Neurol Neurosurg 190:105652. https://doi.org/10.1016/j.clineuro.2019.105652
Siegal T, Charbit H, Paldor I, Zelikovitch B, Canello T, Benis A, Wong ML, Morokoff AP, Kaye AH, Lavon I (2016) Dynamics of circulating hypoxia-mediated miRNAs and tumor response in patients with high-grade glioma treated with bevacizumab. J Neurosurg 125:1008–1015. https://doi.org/10.3171/2015.8.JNS15437
Molania R, Gagnon-Bartsch JA, Dobrovic A, Speed TP (2019) A new normalization for nanostring nCounter gene expression data. Nucleic Acids Res 47:6073–6083. https://doi.org/10.1093/nar/gkz433
Andersen P, Gill R (1982) Cox's regression model for counting processes: a large sample study. Ann Stat. https://doi.org/10.1214/aos/1176345976
Burgos K, Malenica I, Metpally R, Courtright A, Rakela B, Beach T, Shill H, Adler C, Sabbagh M, Villa S, Tembe W, Craig D, Van Keuren-Jensen K (2014) Profiles of extracellular miRNA in cerebrospinal fluid and serum from patients with Alzheimer's and Parkinson's diseases correlate with disease status and features of pathology. PLoS ONE 9:e94839. https://doi.org/10.1371/journal.pone.0094839
Teplyuk NM, Mollenhauer B, Gabriely G, Giese A, Kim E, Smolsky M, Kim RY, Saria MG, Pastorino S, Kesari S, Krichevsky AM (2012) MicroRNAs in cerebrospinal fluid identify glioblastoma and metastatic brain cancers and reflect disease activity. Neuro Oncol 14:689–700. https://doi.org/10.1093/neuonc/nos074
Stylli SS, Adamides AA, Koldej RM, Luwor RB, Ritchie DS, Ziogas J, Kaye AH (2016) miRNA expression profiling of cerebrospinal fluid in patients with aneurysmal subarachnoid hemorrhage. J Neurosurg. https://doi.org/10.3171/2016.1.JNS151454
Akers JC, Hua W, Li H, Ramakrishnan V, Yang Z, Quan K, Zhu W, Li J, Figueroa J, Hirshman BR, Miller B, Piccioni D, Ringel F, Komotar R, Messer K, Galasko DR, Hochberg F, Mao Y, Carter BS, Chen CC (2017) A cerebrospinal fluid microRNA signature as biomarker for glioblastoma. Oncotarget 8:68769–68779. https://doi.org/10.18632/oncotarget.18332
Drusco A, Bottoni A, Lagana A, Acunzo M, Fassan M, Cascione L, Antenucci A, Kumchala P, Vicentini C, Gardiman MP, Alder H, Carosi MA, Ammirati M, Gherardi S, Luscri M, Carapella C, Zanesi N, Croce CM (2015) A differentially expressed set of microRNAs in cerebro-spinal fluid (CSF) can diagnose CNS malignancies. Oncotarget 6:20829–20839
Bronisz A, Godlewski J, Wallace JA, Merchant AS, Nowicki MO, Mathsyaraja H, Srinivasan R, Trimboli AJ, Martin CK, Li F, Yu L, Fernandez SA, Pecot T, Rosol TJ, Cory S, Hallett M, Park M, Piper MG, Marsh CB, Yee LD, Jimenez RE, Nuovo G, Lawler SE, Chiocca EA, Leone G, Ostrowski MC (2011) Reprogramming of the tumour microenvironment by stromal PTEN-regulated miR-320. Nat Cell Biol 14:159–167. https://doi.org/10.1038/ncb2396
Schepeler T, Reinert JT, Ostenfeld MS, Christensen LL, Silahtaroglu AN, Dyrskjot L, Wiuf C, Sorensen FJ, Kruhoffer M, Laurberg S, Kauppinen S, Orntoft TF, Andersen CL (2008) Diagnostic and prognostic microRNAs in stage II colon cancer. Cancer Res 68:6416–6424. https://doi.org/10.1158/0008-5472.CAN-07-6110
Perez-Carbonell L, Sinicrope FA, Alberts SR, Oberg AL, Balaguer F, Castells A, Boland CR, Goel A (2015) MiR-320e is a novel prognostic biomarker in colorectal cancer. Br J Cancer 113:83–90. https://doi.org/10.1038/bjc.2015.168
Manterola L, Guruceaga E, Gallego Perez-Larraya J, Gonzalez-Huarriz M, Jauregui P, Tejada S, Diez-Valle R, Segura V, Sampron N, Barrena C, Ruiz I, Agirre A, Ayuso A, Rodriguez J, Gonzalez A, Xipell E, Matheu A, Lopez de Munain A, Tunon T, Zazpe I, Garcia-Foncillas J, Paris S, Delattre JY, Alonso MM (2014) A small noncoding RNA signature found in exosomes of GBM patient serum as a diagnostic tool. Neuro Oncol 16:520–527. https://doi.org/10.1093/neuonc/not218
Halle B, Thomassen M, Venkatesan R, Kaimal V, Marcusson EG, Munthe S, Sorensen MD, Aaberg-Jessen C, Jensen SS, Meyer M, Kruse TA, Christiansen H, Schmidt S, Mollenhauer J, Schulz MK, Andersen C, Kristensen BW (2016) Shift of microRNA profile upon orthotopic xenografting of glioblastoma spheroid cultures. J Neurooncol 128:395–404. https://doi.org/10.1007/s11060-016-2125-x
Zheng G, Du L, Yang X, Zhang X, Wang L, Yang Y, Li J, Wang C (2014) Serum microRNA panel as biomarkers for early diagnosis of colorectal adenocarcinoma. Br J Cancer 111:1985–1992. https://doi.org/10.1038/bjc.2014.489
Ogata-Kawata H, Izumiya M, Kurioka D, Honma Y, Yamada Y, Furuta K, Gunji T, Ohta H, Okamoto H, Sonoda H, Watanabe M, Nakagama H, Yokota J, Kohno T, Tsuchiya N (2014) Circulating exosomal microRNAs as biomarkers of colon cancer. PLoS ONE 9:e92921. https://doi.org/10.1371/journal.pone.0092921
Polytarchou C, Oikonomopoulos A, Mahurkar S, Touroutoglou A, Koukos G, Hommes DW, Iliopoulos D (2015) Assessment of circulating microRNAs for the diagnosis and disease activity evaluation in patients with ulcerative colitis by using the nanostring technology. Inflamm Bowel Dis 21:2533–2539. https://doi.org/10.1097/MIB.0000000000000547
Goze C, Reynes C, Forestier L, Sabatier R, Duffau H (2018) Pilot study of whole blood microRNAs as potential tools for diffuse low-grade gliomas detection. Cell Mol Neurobiol 38:715–725. https://doi.org/10.1007/s10571-017-0536-7
Ding Q, Shen L, Nie X, Lu B, Pan X, Su Z, Yan A, Yan R, Zhou Y, Li L, Xu J (2018) MiR-223-3p overexpression inhibits cell proliferation and migration by regulating inflammation-associated cytokines in glioblastomas. Pathol Res Pract 214:1330–1339. https://doi.org/10.1016/j.prp.2018.05.012
Huang BS, Luo QZ, Han Y, Huang D, Tang QP, Wu LX (2017) MiR-223/PAX6 axis regulates glioblastoma stem cell proliferation and the chemo resistance to TMZ via regulating PI3K/Akt pathway. J Cell Biochem 118:3452–3461. https://doi.org/10.1002/jcb.26003
Lan F, Yue X, Xia T (2020) Exosomal microRNA-210 is a potentially non-invasive biomarker for the diagnosis and prognosis of glioma. Oncol Lett 19:1967–1974. https://doi.org/10.3892/ol.2020.11249
Bernardi D, Padoan A, Ballin A, Sartori M, Manara R, Scienza R, Plebani M, Della Puppa A (2012) Serum YKL-40 following resection for cerebral glioblastoma. J Neurooncol 107:299–305. https://doi.org/10.1007/s11060-011-0762-7
Vietheer JM, Rieger J, Wagner M, Senft C, Tichy J, Foerch C (2017) Serum concentrations of glial fibrillary acidic protein (GFAP) do not indicate tumor recurrence in patients with glioblastoma. J Neurooncol 135:193–199. https://doi.org/10.1007/s11060-017-2565-y
Tie J, Kinde I, Wang Y, Wong HL, Roebert J, Christie M, Tacey M, Wong R, Singh M, Karapetis CS, Desai J, Tran B, Strausberg RL, Diaz LA Jr, Papadopoulos N, Kinzler KW, Vogelstein B, Gibbs P (2015) Circulating tumor DNA as an early marker of therapeutic response in patients with metastatic colorectal cancer. Ann Oncol 26:1715–1722. https://doi.org/10.1093/annonc/mdv177
Dawson SJ, Tsui DW, Murtaza M, Biggs H, Rueda OM, Chin SF, Dunning MJ, Gale D, Forshew T, Mahler-Araujo B, Rajan S, Humphray S, Becq J, Halsall D, Wallis M, Bentley D, Caldas C, Rosenfeld N (2013) Analysis of circulating tumor DNA to monitor metastatic breast cancer. N Engl J Med 368:1199–1209. https://doi.org/10.1056/NEJMoa1213261
Lavon I, Refael M, Zelikovitch B, Shalom E, Siegal T (2010) Serum DNA can define tumor-specific genetic and epigenetic markers in gliomas of various grades. Neuro-oncol 12:173–180. https://doi.org/10.1093/neuonc/nop041
Bettegowda C, Sausen M, Leary RJ, Kinde I, Wang Y, Agrawal N, Bartlett BR, Wang H, Luber B, Alani RM, Antonarakis ES, Azad NS, Bardelli A, Brem H, Cameron JL, Lee CC, Fecher LA, Gallia GL, Gibbs P, Le D, Giuntoli RL, Goggins M, Hogarty MD, Holdhoff M, Hong SM, Jiao Y, Juhl HH, Kim JJ, Siravegna G, Laheru DA, Lauricella C, Lim M, Lipson EJ, Marie SK, Netto GJ, Oliner KS, Olivi A, Olsson L, Riggins GJ, Sartore-Bianchi A, Schmidt K, Shih M, Oba-Shinjo SM, Siena S, Theodorescu D, Tie J, Harkins TT, Veronese S, Wang TL, Weingart JD, Wolfgang CL, Wood LD, Xing D, Hruban RH, Wu J, Allen PJ, Schmidt CM, Choti MA, Velculescu VE, Kinzler KW, Vogelstein B, Papadopoulos N, Diaz LA Jr (2014) Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med 6:ra224. https://doi.org/10.1126/scitranslmed.3007094
Miller AM, Shah RH, Pentsova EI, Pourmaleki M, Briggs S, Distefano N, Zheng Y, Skakodub A, Mehta SA, Campos C, Hsieh WY, Selcuklu SD, Ling L, Meng F, Jing X, Samoila A, Bale TA, Tsui DWY, Grommes C, Viale A, Souweidane MM, Tabar V, Brennan CW, Reiner AS, Rosenblum M, Panageas KS, DeAngelis LM, Young RJ, Berger MF, Mellinghoff IK (2019) Tracking tumour evolution in glioma through liquid biopsies of cerebrospinal fluid. Nature 565:654–658. https://doi.org/10.1038/s41586-019-0882-3
Murray MJ, Watson HL, Ward D, Bailey S, Ferraresso M, Nicholson JC, Gnanapragasam VJ, Thomas B, Scarpini CG, Coleman N (2018) "Future-proofing" blood processing for measurement of circulating miRNAs in samples from biobanks and prospective clinical trials. Cancer Epidemiol Biomark Prev 27:208–218. https://doi.org/10.1158/1055-9965.EPI-17-0657
Marzi MJ, Montani F, Carletti RM, Dezi F, Dama E, Bonizzi G, Sandri MT, Rampinelli C, Bellomi M, Maisonneuve P, Spaggiari L, Veronesi G, Bianchi F, Di Fiore PP, Nicassio F (2016) Optimization and standardization of circulating microRNA detection for clinical application: the miR-test case. Clin Chem 62:743–754. https://doi.org/10.1373/clinchem.2015.251942
Acknowledgements
We would like to thank Professor Mark Rosenthal for helpful discussions and comments regarding this work.
Funding
Cure Brain Cancer Foundation Australia, Brain Foundation Australia, The Royal Melbourne Hospital Neuroscience Foundation.
Author information
Authors and Affiliations
Contributions
Conception, planning: AM, AK, TS, KD. Writing & editing manuscript: AM, JJ, HN, KD, TS, FG, AK. Experimental work: HN, JJ, IB, RL, SS, RK, IP, LP, CM. Biostatistics: JL, AW, GI, CM, TS, RM, TK, CM. Radiology analysis: FG, AL, JJ.
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare no conflict of interest.
Ethics approval
Ethics approval for this study was obtained from the Melbourne Health Human Research Ethics Committee (HREC 2009.114), Melbourne, Australia.
Informed consent
All patients provided written informed consent prior to inclusion. The study was performed in accordance with the Declaration of Helsinki.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Morokoff, A., Jones, J., Nguyen, H. et al. Serum microRNA is a biomarker for post-operative monitoring in glioma. J Neurooncol 149, 391–400 (2020). https://doi.org/10.1007/s11060-020-03566-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11060-020-03566-w