Abstract
Purpose
The woolly beech aphid thrives on European beech leaves, which has complex direct and indirect impacts on above- and belowground processes. A mechanistic understanding of insect-mediated changes in organic carbon (OC) availability for microbial life and its implications for element cycling is still lacking. This study aims at disentangling aphid-induced effects on phyllosphere and rhizosphere bacterial communities, as well as investigating feedbacks to OC transfer from the canopy to the mineral soil.
Methods
Following 2.5 months of infestation, we tracked the fate of OC (13CO2 pulse-labelling) in several compartments of beech sapling – soil mesocosms over 5 days. In ecosystem solutions, water extracts and soil/plant compartments we determined OC and N and solid δ13C. Bacterial community structure (16S rRNA gene targeted amplicon sequencing and quantitative PCR) and metabolite profiles (LC-qTOF-MS) were analysed.
Results
We found significantly higher aphid-mediated inputs of OC within throughfall. Honeydew-derived C on infested leaves was inconsequential for total phyllosphere bacterial abundances, but verifiably affected the community structure. In all soil compartments, cold-water extractable OC pools declined significantly by frequent inputs of readily available OC. This pattern might relate to reductions in rhizodepositions and altered microbial processing by accelerated soil C-mineralization. As a result, the abundance of metabolites changed significantly in different ecosystem solutions.
Conclusions
Our findings attest that insect infestations induce distinct direct and indirect effects on plant-insect-microbiome interactions leading to marked alterations in C dynamics. This integrated approach improves our understanding on microbial dynamics and biogeochemistry and evaluates the role of insects for ecosystem processes.
Similar content being viewed by others
Introduction
In forest ecosystems, pest outbreaks of phytophagous insects can directly and indirectly affect a wide spectrum of key ecosystem processes and functions, such as primary productivity of plants and the turnover of organic matter (Schowalter 2016). Direct effects reduce the plant’s ability to assimilate and internally translocate photosynthates to carbon (C) sinks like roots. In addition, sap-feeding insects, directly affect belowground biota via their excrements, such as frass and honeydew, entering the forest floor. Indirect effects are the result of systemic induced responses in their host plants (van Dam and Heil 2011). Both direct and indirect effects lead to complex interactions between aboveground and belowground biota, which may affect ecosystem organic matter (OM) and nutrient cycling to different degrees and directions (Bardgett and Wardle 2010).
The plant’s ability to assimilate C can be rapidly reduced by sap-feeding insects such as aphids. For instance, the amount of photo-assimilates diverted from phloem by sap-feeders may account for 6–8% of the net primary productivity (Beggs et al. 2005). Furthermore, C and N allocation patterns in plants are directly affected by insect herbivory changing the amount and composition of rhizodepositions and, thus, altering the resource availability for soil biota (Bardgett and Wardle 2010; van Dam and Bouwmeester 2016). Herbivory by chewing insects temporarily increases C allocation from shoots to roots as well as root exudation (Holland et al. 1996; Potthast et al. 2021), whereas aphid feeding triggers the opposite effect (Smith and Schowalter 2001; Vestergård et al. 2004). We therefore suppose that aphid infestation decelerates the carbohydrate release to mycorrhizae and/or rhizosphere organisms (Gange et al. 2002; Vestergård et al. 2004).
Aphid feeding does not only change the quantity but also the quality of the rhizodeposition, which has shown changes in the composition (Kim et al. 2016; Vestergård et al. 2004) and microbial richness of the rhizosphere community (Liu et al. 2020). For instance, Gram-positive bacteria (Paenibacillus spp.) were enhanced in the rhizosphere of pepper (Capsicum annuum) infested by sap sucking insects (Kim et al. 2016), which affected the resistance of their host plant towards further herbivory or pathogen attacks (Kim et al. 2016; Yang et al. 2011).
Aphids excrete most of the phloem-derived photo-assimilates as sugars that often crystalize as so called “honeydew” in large droplets on leaf surfaces. Depending on the aphid and its host plant species, an individual aphid may excrete between ~4 and 2000 μg honeydew h−1 (Moir et al. 2018). In forested ecosystems, the amounts of honeydew produced may yield 400–700 kg fresh weight per hectare and year (Zoebelein 1954). These sugars, mainly fructose, glucose and melezitose (Douglas 2006), may become directly available as energy source for phyllosphere microorganisms (Stadler and Müller 2000). In the phyllosphere, different groups of organisms such as yeasts, bacteria, filamentous fungi, and epiphytic plants (Kinkel 1997) contribute to OM transformation and decay. They are likely to cause nutrient immobilization, thereby changing the biochemical composition of solutions in the ecosystem (Michalzik et al. 2016). For example, in the presence of easily available C resources, phyllosphere microorganisms can grow and immobilize inorganic N. This may cause reduced fluxes of inorganic N during peak insect feeding activity (Stadler et al. 2001).
Direct inputs of large quantities of easily available honeydew to organic layer and topsoils likely leads to the acceleration of soil mineralization rates (Michalzik and Stadler 2005) which, in turn, will affect soil OM and nutrient dynamics (Choudhury 1985). Dissolved OM levels in soil solutions and extracts are sensitive indicators for changes in such dynamics (Zsolnay 2003). For instance, cold-water soil extracts reflect nutrient and energy sources for soil microbes (Jandl and Sollins 1997) and contain other metabolites resulting from belowground plant and microbial processes (Gregorich et al. 2003; Landsman-Gerjoi et al. 2020). Impacts of aphid infestation on easily available OM and on secondary metabolites likely reflect changes in rhizodeposition and/or an altered microbial processing of available substrate, in particular, in the organic layer as an important part of the total soil OM pool.
Following this line of argumentation, we expected aphid infestation to affect microorganisms both in the phyllos- and in the rhizosphere via direct (honeydew) and indirect effects (plant internal responses) with consequences for key ecosystem processes such as C and N cycling. To gain a better mechanistic understanding of these aphid-mediated processes, we used a mesocosm set-up with 13CO2 pulse-labelling in the main infestation phase to experimentally assess the effect of a woolly aphid (Phyllaphis fagi) infestation on four-year-old beech saplings. In this context, we hypothesized that honeydew excretion will 1) increase the microbial abundance and diversity in the phyllosphere due to higher C availability, and 2) will increase the availability of OM resources in the soil via the input of leached honeydew sugar from leaves. We further assumed that 3) aphid-mediated higher C input to the forest floor alters metabolite profiles of ecosystem solutions and extracts (throughfall, forest floor, root zone, mineral soil) collected from aphid and control treatments, and that 4) aphid attack will reduce rhizosphere bacterial diversity and abundance due to reduced plant-C allocations which may suppress copiotrophic bacteria. To better understand the underlying mechanisms, we pursued an integrated approach, linking metabolomics and microbial community analyses with elemental tracing (13C) and Scanning Electron Microscopy (SEM) imaging.
Material and methods
Experimental set-up
We designed a mesocosm experiment with European beech (F. sylvatica L.) saplings to standardize our investigations with regard to tree age, nutritional stage, irrigation volumes and controlling of aphid infestation. In brief, our experiment consisted of three phases: the preparation (3 months) and aphid infestation (2.5 months) phase outdoors and the subsequent five-day incubation phase in mesocosms under controlled conditions in a climatic chamber. A detailed description of the set-up of our custom-made mesocosms (196 l volume) is provided by Potthast et al. (2021). Table 1 elucidates the experimental timeline. The preparation phase encompassed the collection of organic layer (F-Mull) and top soil material (stagnic Luvisols) from a European beech dominated forest of the Hainich Critical Zone Exploratory (CZE), which was launched as an interdisciplinary research platform within the Collaborative Research Center (CRC) AquaDiva (Küsel et al. 2016). Two four-year old beech saplings each (60–70 cm in height) were potted in homogenized top soil in the bottom parts of 12 mesocosms (Fig. 1 left). After planting, the Oi and Oe horizons of the organic layer were carefully placed on top of the mineral soil following the natural dimensions as observed at the forest site.
Experimental set-up – Left: non-infested beech saplings equipped with soil moisture and temperature probe as well as with a glass and a plastic funnel for throughfall and organic layer leachate sampling, respectively. Right: Four gas-tight mesocosms (Control; Labelled (+13CO2); Aphid (+13CO2 + moderately infested); Aphid heavy (+13CO2 + heavily infested)) infested for 2.5 months and afterwards incubated for 5 days in a climatic chamber
At the beginning of May, aphid infestation started to emerge in the field. Half of the mesocosms (n = 6) were spatially separated (50 m apart, separated by a wall of an old greenhouse without roof) to avoid natural infestation of control treatments. The other half was artificially infested with 20 individuals of the woolly beech aphid (Phyllaphis fagi L.) per mesocosm to mimic field infestation patterns. Early July, one boro-silicate glass suction plate (ecoTech, Bonn, diameter 8 cm) per mesocosm was placed in the root zone in 4 cm soil depth. In late July, we started to prepare the mesocosms for the incubation experiment. We randomly placed one throughfall sampler (glass funnel, diameter 12 cm) and one forest floor lysimeter (PE funnels, diameter 12 cm, filled with Oi and Oe material) underneath each canopy (see Fig. 1 left) to measure the input to the forest floor and the output as organic layer leachates into the mineral soil. Mesocosm output fluxes were collected by seepage losses at the outlet port of the bottom plate (Fig. 1 right). Soil moisture and temperature probes were installed in the top mineral soil to adjust similar soil moisture conditions in all mesocosms (Fig. 1) and to allow constant monitoring during incubation.
For the aphid treatments, differences in emerging aphid infestation levels were observed from early to mid-June onwards. Aphids were the most abundant in mid-July still exhibiting diverting infestation levels. Accordingly, two infestation levels were defined for the incubation experiment with half (n = 3) of the mesocosms showing a heavy infestation level with nearly all twigs infested by aphids and > 30 aphids above throughfall and forest floor samplers and half (n = 3) with moderate levels with <50% of the twigs infested and with 10–20 individuals above the samplers. As a result, we distinguished four treatment groups with three replicates each for the incubation experiment: C = 12CO2 (Control), L = 13CO2 without aphid infestation (Labelled), A = 13CO2 + moderate aphid infestation (Aphid), AH = 13CO2 + high aphid infestation (Aphid heavy).
Incubation phase -13CO2 pulse-labelling
Before the start of the five-day incubation experiment, mesocosms were equipped with two ventilators for headspace gas mixing and sealed gas-tight (Fig. 1 right). Afterwards, mesocosms were flushed with C-free air followed by a stepwise release of CO2 after addition of HCl to CaCO3 (6.4242 mmol CO2 for the Control and 13CO2 for the labelled mesocosms). A detailed description of the 13CO2 pulse-labelling procedure is provided by Potthast et al. (2021). During incubation, mesocosms were irrigated with 3.6 l tap water per mesocosm (corresponding to 18.3 mm) by applying 400, 600, 600, 2000 ml after 48, 80, 89, 120 h, respectively. Event-based throughfall and soil solution volumes were documented and samples were pooled for further analyses. Lysimeter seepage solutions were cumulatively sampled once at the end of the experiment following the last precipitation event. The total volume of irrigation subtracted by the respective collected volumes of throughfall and of OM solution (3.6 l cosm−1 – sample volumes) was multiplied with element concentrations to calculate input budgets for each mesocosm.
After terminating the incubation, beech leaf material for phyllosphere microbiome analyses was picked from all treatments and transferred to autoclaved 0.5 L polypropylene ether (PPE) containers (see 2.4). The subsequent destructive sampling encompassed the quantitative collection of Oi and Oe layer material, the whole beech tree harvest by separating leaves and above-ground stem and below-ground root biomass. The soil adhering directly to the beech roots to a maximum of one centimetre was defined as the rhizosphere soil (Tischer et al. 2019). For rhizosphere microbiome analysis, about 2 g of soil were obtained from five different locations within the rhizosphere using a sterile spatula, transferred to sterile reaction tubes, frozen immediately on dry ice, and stored at −80 °C. After rhizosphere sampling, roots were washed with tap water. Mineral soil samples were obtained by three soil cores (steel cylinder, height 12 cm, diameter 9.5 cm) per mesocosm and pooled for further analysis.
Scanning electron microscopy (SEM)
To visualise the impact of woolly aphids on the surface conditions of the leaf undersides, one leaf sample from each treatment was harvested during destructive sampling and prepared for SEM analysis. Upon collection, randomly selected portions of fresh leaf samples were mounted to 10 mm diameter SEM discs. Dried discs, according to the protocol of Pathan et al. (2008), were each sputtered with 20 nm gold for SEM (Zeiss Auriga 60 with field emission cathode and Everhart-Thornley type SE detector, Carl Zeiss SMT, Oberkochen, Germany). The scanning electron microscope has a resolution of 1.0 nm at 15 kV. Each individual disc was imaged at magnifications of 20x and 100x.
Chemical analyses
Throughfall, organic layer and soil solutions were vacuum-filtered to <0.45 μm with prewashed membrane filters (cellulose acetate, Sartorius, Göttingen, Germany) and analysed for dissolved organic C (DOC) and total dissolved N (TDN) by thermal oxidation (TOC-VCPN/TNM-1, Shimadzu, Duisburg, Germany). Solution pH value (WTW pH-Elektrode SenTix81, Weilheim, Germany) and electrical conductivity (WTW TetraCon 325, Weilheim, Germany) were analysed as well.
Immediately after destructive sampling, water-extractable organic matter was obtained by cold-water extraction in triplicate from 10 g of field fresh rhizosphere or mineral soil samples and from 3 g of fresh Oi and Oe layer samples, respectively, weighted into Falcon-tubes. Thirty ml of distilled water were added, samples were shaken on a horizontal shaker for one hour (150 r/min), centrifuged for 5 min at 2800 r/min and vacuum-filtered to <0.45 μm as above. Concentrations of DOC and TDN in solutions were analysed, then normalized to the used soil amount and water volume and expressed as mg g−1 dry soil.
Leaves, roots and organic layer samples (Oi, Oe) were freeze-dried, whereas soil samples (rhizosphere and mineral soil) were dried at 40 °C until constant mass. All samples were ground and measured for organic C (TOC) and total N (TN) using an elemental analyzer (Elementar Vario El Cube; Elementar Analysensysteme GmbH, Germany) and for δ13C using an EA-IRMS (CE 1100 coupled via Con Flo III with a Delta+; Thermo Fischer, Germany).
Microbiome community structure analyses
Preparation of samples for analysis of phyllosphere bacterial communities was carried out as described previously (Herrmann et al. 2021). In brief, 250 ml of suspension buffer (0.15 M NaCl, 0.1% Tween 20) were added to the containers in which leaves had been sampled, followed by mild sonication (2 × 1 min at 10% intensity), shaking at 100 r/min at room temperature for 20 min, and filtration of suspensions on 0.2 μm polyethersulfone filters (Supor, Pall Corporation) using a vacuum device. Filters were stored at −80 °C until nucleic acid extraction. Leaf material was dried at 50 °C for one week for determination of dry weight.
Extraction of DNA from phyllosphere and rhizosphere samples was performed using the DNEasy PowerSoil kit (Qiagen, Germany) following the manufacturer’s instructions. Filter material from half a filter cut into small pieces or 0.2 g of soil were used as starting material.
Quantification of bacterial 16S rRNA genes in rhizosphere soil samples was carried out by quantitative PCR using Brilliant II SYBR Green Mastermix (Agilent Technologies) on a Mx3000P qPCR cycler (Agilent Technologies) with the primer combination Bac8Fmod/Bac338Rabc (Loy et al. 2002; Nercessian et al. 2005) following the cycling conditions described in Herrmann et al. (2012). To avoid amplification of chloroplast-derived 16S rRNA genes, we used a different primer combination for the phyllosphere samples - Bakt_0341F (Klindworth et al. 2013) and Bakt_0799R, the reverse complement of primer 799F (Chelius and Triplett 2001) with cycling conditions as in Herrmann et al. (2021).
Amplicon sequencing of bacterial 16S rRNA genes was performed using the primer combination Bakt_0341F/Bakt_0785R (Klindworth et al. 2013) and the Illumina MiSeq platform (v3 chemistry) at LGC Genomics (phyllosphere samples) and in house sequencing (rhizosphere samples) as previously described (Krüger et al. 2021; Rughöft et al. 2016).
Sequences were analysed using Mothur (Schloss et al. 2009) following the Mothur SOP (Kozich et al. 2013) along with the SILVA v132 references for alignment and taxonomic classification (Quast et al. 2013). Chimera check was performed using the ‘uchime’ algorithm implemented in Mothur, and Operational Taxonomic Units (OTUs) were assigned using ‘vsearch’ in Mothur with a 0.03 distance cut-off. Shannon diversity indices and estimated numbers of OTUs (Chao) were derived within the Mothur pipeline. Sequence data generated in this study have been submitted to the European Nucleotide Archive (ENA) under the study accession number PRJEB46752, sample accession numbers SAMEA9456885 – SAMEA9456904 and SAMEA10017471-SAMEA10017482.
Estimation of cell abundances
Taxonomic information was combined with results of qPCR analyses to estimate the number of cells in phyllosphere and rhizosphere samples. To do so, we calculated relative abundances of each OTU present in these samples. Following the OTU-based taxonomic assignment from the Mothur pipeline, we retrieved information from the rrnDB database (https://rrndb.umms.med.umich.edu, accessed April 2020 (Stoddard et al. 2014) how many 16S rRNA operons are on average present in the corresponding taxa (genus or family level where available). Relative abundances of each OTU were then multiplied with 16S rRNA gene abundances of the respective sample and corrected for multiple 16S rRNA operons where necessary.
Liquid chromatography metabolite profiling
An aliquot of ecosystem solutions and cold-water extracts was freeze dried and kept at −80 °C before further clean up (Delory et al. 2021). The sample residue was re-constituted with 2 ml of ultrapure water (MilliQ) containing 5% Methanol (v/v%), sonicated for 10 min at room temperature and transferred to a new safe lock tube. Afterwards, samples were centrifuged for 10 min at 6000 g and 4 °C and the supernatant was transferred to a new tube for clean up by solid phase extraction (SPE). One millilitre of a sample aliquot was loaded on the column to clean it up on a SPE column (Chromabond C18-Hydra 200 mg/3 ml Macherey Nagel), that was previously conditioned with 2 ml of methanol (LC-MS grade, VWR) followed by 2 ml of ultrapure water containing 2% formic acid (v/v%) (LC-MS grade, Sigma Aldrich). After the column was washed with 2 ml of ultrapure water, samples were eluted with 2 ml methanol containing 2% formic acid (v/v%) (LCMS grade, VWR). The eluate was evaporated to dryness in a vacuum concentrator at 40 °C and reconstituted in 150 μl of methanol containing 30% water (v/v%). The samples were sonicated for 10 min at room temperature and then centrifuged at 6000 g and 4 °C for 10 min. The supernatant was then transferred to 1.5 ml LC vials.
Metabolite profiling was done on a Bruker Maxis impact HD qTOF-MS coupled to a Dionex 3000 liquid chromatography system. More detailed information about the applied procedure can be found in the supplementary material.
Processing of raw LC-MS data
The LC-qToF-MS raw data were converted to the mzXML format using the CompassXport utility of the Data Analysis software (Bruker Daltonics). Peak picking, feature alignment and feature grouping was done in R 3.6.0 (R core team 2019) using the Bioconductor (Huber et al. 2015) packages ‘xcms’ v1.52.0 (Tautenhahn et al. 2008) and ‘CAMERA’ v1.32.0 (Kuhl et al. 2012). For raw data pre-processing with ‘xcms’ and ‘CAMERA’, all samples were organized into four groups, according to their treatments: Control, Labelled, Aphid, Aphid heavy. The following ‘xcms’ parameters were applied: peak picking method ‘centWave’ (snthr = 10; ppm = 10; peakwidth = 2, 10); peak grouping method ‘density’ (minfrac = 0.7; bw = 3; mzwid = 0.05); retention time correction method ‘peakgroups’ (family = symmetric). CAMERA was used to annotate adducts, fragments, and isotope peaks with the following parameters: extended rule set (https://github.com/stanstrup/commonMZ/tree/master/inst/extdata); perfwhm = 0.6; calcIso = TRUE; calcCaS = TRUE. CAMERA additionally sorted these adducts/ fragments into pseudo compound (PC) groups where each group potentially represents a metabolite. Lastly, each PC group (referred to as ‘metabolite’) was collapsed using an in-house maximum heuristic approach aiming to find the feature that most often displayed the highest intensity values across all samples for each PC group (Ristok et al. 2019). In our analysis, each PC group was therefore represented by one feature with a known mass-to-charge ratio (mz) and retention time (rt).
Statistical analyses
All response variables of plant, soil, and solution properties were described by mean and standard error (SE) or standard deviation (SD) as derived from the statistical models. Data were checked for normality and homogeneity of model residuals, and, if necessary, were transformed using log10. Effects were analysed with linear mixed effect models (LMMs) (Pinheiro et al. 2013) in the R statistical environment, R 3.5.0 (R core team 2019) using contributed package nlme (Pinheiro et al. 2013).
As fixed factor, we included treatment and type. We considered all respective treatments (C, L, A, AH) as treatment levels. Depending on the tested ecosystem compartments, type was specified for biomass compartments, for solution type, and for soil, respectively. Biomass compartments were separated into aboveground and belowground. Solution type was separated into throughfall, suction cups, forest floor and bottom lysimeters. Soil was separated into bulk and rhizosphere soil. Due to non-linearity in response variables, mineral soil and organic layer were separated for TOC and TN analysis. Mesocosm ID was set as random effect to account for nested sampling of several biomass compartments, solution types, and soil strata within the same mesocosm. To allow for different variances for each level of type and hence to account for heteroscedasticity of the model, we finally used the varIdent class of the varFunc argument (Pinheiro et al. 2013). The concentrations, isotopic signatures, and stocks of elements in biomass, solution, soil and cold-water extracts of soil were included as response variable in the LMMs, respectively.
The final LMMs included 2-way interactions of treatment and type. These “full” models were used for model parameter estimation unless 2-way interactions were significant or not (Schielzeth and Forstmeier 2009). The final model includes variance heterogeneity among treatments or types, depending on the performance of the model in terms of Akaike Information Criteria (AIC, see below) and patterns in model residuals. The procedure of model building and final model selection was equal as described in Potthast et al. (2021). The final model estimates were reported in the supplement (Tables S3–7) and are based on Restricted maximum likelihood (REML) estimates. Post-hoc tests were performed using emmeans package in R and estimates were adjusted for false discovery rate (Verhoeven et al. 2005).
Differences in abundances of bacterial 16S rRNA genes or relative abundances of bacterial taxonomic groups between samples were analysed using pairwise Mann-Whitney tests. Principal Coordinate Analysis (PCoA) of samples based on their bacterial community composition was done using the software PAST (Hammer et al. 2001).
We used PERMANOVA analysis in R (R core team 2019) to test for the effect of treatments on phyllosphere and rhizosphere bacterial community composition, using the Adonis functions from the R package vegan (Oksanen et al. 2015) and respective dependencies.
LCMS data were exported to Metaboanalyst 4.0 (Chong et al. 2019). A principal component analysis (PCA) was applied after normalization of data to the amount of DOC and Pareto scaling. Hierarchical Clustering was done with Euclidean distances and Ward’s method as clustering algorithm. Chemical richness was calculated as the sum of all metabolites and chemical diversity as the Shannon diversity of all metabolites using the R-package vegan (Oksanen et al. 2015).
Results
Plant and soil properties
According to LMM results, basic plant and soil properties were not significantly different between treatments (Table S3, S5, S6). Briefly, total beech biomass ranged from 55.6 to 60.3 g dry matter (DM) per mesocosm for aboveground biomass and from 21.4 to 28.6 g DM for roots. The C:N ratios of leaf and root biomass ranged from 26 to 29 and 31–35, respectively. The amount of OC in the Oi layer was 1.6 times higher than in the Oe layer with C:N ratios ranging from 28 to 32 and from 21 to 23 for the Oe layer. SOC and TN amounts of bulk and rhizosphere soil extended between 1.7–1.8% and between 0.12–0.13%, respectively (Table S1).
The abundance of 13C in above- and belowground biomass increased significantly five days after 13CO2-amendment (p < 0.0001) (Fig. 2a). The highest 13C abundances were found in leaves followed by roots > Oi layer > rhizosphere soil > bulk mineral soil > Oe layer. Compared to the control, roots were significantly enriched in 13C in the labelled treatment, but not in any of the aphid treatments (Fig. 2a). The latter showed significantly higher δ13C values only in the Oi layer compared to the labelled treatment (Fig. 2b), but not for the Oe layer and the bulk mineral soil. LMM results revealed significantly higher δ13C values in rhizosphere soil compared to bulk mineral soil (p = 0.0181), which was mainly associated with 13CO2 amendment (p = 0.0635). The treatments did not significantly differ in bulk mineral soil 13C abundance (Fig. 2b), whereas δ13C values in rhizosphere soil were higher in the labelled treatments than in the Control (L: p = 0.0855; A: p = 0.0056; AH: p = 0.0599, Table S6).
Abundance of 13C in leaves and roots (a) and in organic layer (Oi, Oe) and mineral soil (bulk, rhizosphere) (b) depending on treatment: Control (+12CO2), Labelled (+13CO2), Aphid (+13CO2 + moderately infested), Aphid heavy (+13CO2 + heavily infested). Different lower-case letters indicate significant differences between treatments of each compartment according to LMM (p < 0.05)
Chemistry of soil solutions and cold-water extracts
We found strong effects of treatment (LMM, p = 0.0077) and solution types (p < 0.0001) for DOC concentrations (Table S7). Aphid treatments significantly increased DOC concentrations in throughfall solutions, with the highest levels found in the heavily infested mesocosms (Fig. 3). Whereas leachates of the organic layer and soil solution from 4 cm soil depth did not differ in DOC concentrations, significantly higher values were detected in seepage solutions of the heavy aphid infested mesocosms than in the Control (Fig. 3, Table S7). In contrast, no significant treatment effect (p = 0.2977) became notable for TDN concentrations in any of the solution types (Table S2). Mean TDN concentrations in throughfall ranged from 1.0 to 1.3 mg L−1, which was significantly lower than in other solution types (p < 0.0001). Highest TDN concentrations (mean range: 9.2–23.8 mg L−1) were found in soil solutions collected by suction plates.
Concentrations of dissolved organic carbon (DOC) of different solution types (throughfall, organic layer leachate, soil solutions via suction plate in 4 cm mineral soil depth and via seepage loss) depending on treatment: Control, Labelled, Aphid (moderately infested), Aphid heavy (heavily infested). Different lower-case letters indicate significant differences between treatments of each solution type according to LMM (p < 0.05)
The DOC and TDN amounts (mg g−1 DM) in cold-water extracts showed significant effects of treatment (p < 0.0001) and of different soil compartments (p < 0.0001) (Table S4). Significantly lower amounts of DOC were extracted in all compartments from aphid mesocosms compared to Control (p < 0.0001) and Labelled (p < 0.01) treatments (Fig. 4a). Independent of infestation levels, all aphid treatments showed on average 3.6 and 6.6 times lower water extractable DOC amounts across all compartments compared to Labelled and Control, respectively. Hence, 13CO2 pulse-labelling seemed to additionally affect extractable DOC amounts. The TDN amounts in all compartment solutions were significantly higher in the Labelled than in any other treatment (Fig. 4b) (Table S7). Herbivory also affected C to N ratios in soil extracts (p < 0.0001) with significantly lower ratios across all compartments of the aphid treatments compared to the Control.
Amounts of cold-water extracted dissolved organic carbon DOC (a) and total dissolved nitrogen (TDN) (b) of organic layer (Oi, Oe) and mineral soil (bulk, rhizosphere) depending on treatment: Control, Labelled, Aphid (moderately infested), Aphid heavy (heavily infested). Different lower-case letters indicate significant differences between treatments of each compartment according to LMM p < 0.05)
Microbial phyllosphere and rhizosphere abundance and community composition
Estimation of bacterial cell abundances based on 16S rRNA analyses and taxonomic information from amplicon sequencing revealed that phyllosphere bacterial abundances did not increase over the 2.5 months infestation period. On the contrary, we observed lower abundances in the aphid treatments (estimated cells per g (dw) leaves: 2.86 × 107 (A) and 1.83 × 107 (AH)) compared to the control treatments (5.76 × 107 (C) and 1.49 × 108 (L)) (Fig. S1a). However, these differences were not statistically significant.
Effects of experimental aphid infestation were more pronounced for the bacterial communities in the phyllosphere than for the rhizosphere communities, explaining 24% of the leaf bacterial community variation (Fig. 5a; PERMANOVA, p = 0.012, Table S8). Community shifts were primarily linked to an increase in the relative abundance of Gammaproteobacteria and a decrease in the relative abundance of Actinobacteria and Alphaproteobacteria (Fig. S2). In turn, within the Actinobacteria, the strongest decrease in estimated abundance was observed for the genera Microbacterium and Curtobacterium (Micrococcales), Friedmaniella (Propionibacteriales), Kineococcus (Kineospirales), and Rhodococcus (Corynebacteriales), and for Phenylobacterium (Caulobacterales), Novosphingobium and Sphingomonas (Sphingomonadales) within the Alphaproteobacteria (Fig. S3). Shannon diversity index of the phyllosphere communities did not show significant differences between the aphid and control treatments (Fig. S1b).
Principal Coordinate Analysis of the phyllosphere (a) and rhizosphere (b) bacterial communities based on Bray-Curtis distances. Colours denote origin of samples from the Control (C), Labelled (L), and the two aphid treatments (A: moderately infested, AH: heavily infested). Analysis is based on estimated cell abundances of species-level bacterial taxa
In the next step, we tested the relationship between OTU estimated abundances and the amount of total canopy-derived DOC as a proxy of the amount of DOC available to the phyllosphere microbiota. In total, 51 out of the 1054 OTUs that occurred in the phyllosphere bacterial communities were negatively correlated with the total canopy-derived DOC whereas only two low-abundant unclassified OTUs showed a positive correlation. The negatively correlated OTUs were mostly affiliated with Hymenobacteraceae, Microbacteraceae, Sphingobacteraceae, Sphingomonadaceae, Xanthomonadaceae, and Pseudomonadaceae (Fig. S4).
In the rhizosphere, estimated bacterial cell abundances remained at comparable levels in the control treatments and under the influence of aphid infestation (mean values ranging from 2.41 × 109 to 3.89 × 109 estimated cells per g (dw) soil) (Fig. S1c). However, aphid infestation had a significant effect on bacterial community composition, explaining 8% of the overall variation (Fig. 5b; p = 0.003). In contrast to the phyllosphere communities, variation of the rhizosphere communities was not linked to shifts of major groups on the class or phylum level (Fig. S5) but resulted from changes at lower taxonomic levels. The Shannon diversity index of rhizosphere communities was lower in the aphid treatments compared to the controls but this difference was only significant for the moderate aphid treatment (Fig. S1d). The majority of OTUs which were missing in the aphid treatments were affiliated with Acidobacteria, Alphaproteobacteria, Actinobacteria, and Gammaproteobacteria (Fig. S6). However, most of these OTUs were only present at low abundances in the rhizosphere soil of the control treatments, suggesting that the loss of bacterial taxa under the aphid infestation did not include quantitatively important members of the rhizosphere bacterial community.
Scanning electron microscope imaging
The undersides of aphid-infested beech leaves showed dense accumulations of structures (Fig. 6) that can be assigned to waxy secretions (wax tufts, threads, thick wax bundles). These wax threads extruded from the body of Phyllaphis fagi L. are formed to long hollow skeins where residues of them can be seen on aphid treated leaves. Rod shaped bacteria seemed to be high in densities on the Control and Labelled leaves whereas on the aphid infested leaves these shapes cannot be found in such densities.
Scanning electron microscope images of beech leaf undersides (Fagus sylvatica) of 1 μm (above) and corresponding zoom to 10 μm (below) of different treatments at the end of the experiment. Following treatments were implemented: Control (+12CO2), Labelled (+13CO2), Aphid (Phyllaphis fagi, +13CO2 + moderately infested) and Aphid heavy (+13CO2 + heavily infested)
Metabolomic-profiling
The metabolomite composition of the cold-water extracts and ecosystem solutions from different layers were affected by aphid treatment. Results of a PCA of metabolic profiles showed a significant effect of aphid infestation in the cold-water extracts from the litter layer to the mineral soil (Fig. 7). There, the metabolite profiles of moderate and heavily infested treatments clearly separated along PC1 from the Control and Labelled treatments. Differences of metabolomite composition in the throughfall solutions between treatments were not significant. The chemical diversity and chemical richness, as a proxy of changes in the metabolome, did not significantly change in any of the ecosystem solutions (Fig. 7 and Fig. S7).
Results of metabolomics data analyses of throughfall solutions and of cold-water extracts from litter and fermented organic layer and from mineral and rhizosphere soil normalized by DOC amounts. (a) Principal component analysis of Control (+12CO2 open circle), Labelled (+13CO2 grey circle), moderate (+13CO2 orange circle) and heavy aphid infestation (+13CO2 red circle) p values show results of MRPP, and heat maps (PC groups, Euclidean distance, Ward algorithm) of metabolites. (b) Chemical richness (numbers) and diversity (Shannon) of metabolites (Statistics for 1-Way ANOVA)
Discussion
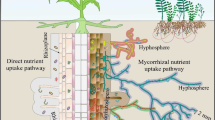
Our results attest that aphids in particular affected the belowground organic carbon pools by both direct and indirect effects. We observed that the direct effects of honeydew excretion on phyllosphere bacterial community were less pronounced. In contrast, we found that more than two months of woolly aphid infestation significantly decreased the water-extractable organic carbon pool in the organic layer, and in bulk and rhizosphere soil. Moderate to high aphid infestation decreased the amounts of extractable OC for soil microbes in all soil compartments whereas the amount of nitrogen remained constant. Indirect mechanisms via plant responses in concert with direct ones via the input of honeydew appear to affect belowground patterns (Fig. 8). In the following, we discuss the contribution of possible direct and indirect effects to the overall mechanism.
Direct effects on above- and belowground biochemical properties
Sap feeders contribute to a continuous input of energy-rich C compounds like fructose, glucose and melizitose to the phyllosphere and as readily available DOC to the organic layer and topsoil (Fischer et al. 2005; Stadler et al. 2001). In this study, the woolly beech aphid caused significantly higher DOC concentrations in throughfall with highest ones in the heavily infested treatment. The majority of these high loads of energy-rich substrates were temporarily stored on the leaves until leaching. In this context, significantly higher population densities (CFUs) of epiphytic filamentous fungi, yeast and bacteria of aphid infested canopies of beech, oak and spruce were reported in the literature (Mühlenberg and Stadler 2005; Stadler and Müller 2000). However, in contrast to our first hypothesis, woolly beech aphid infestation did not result in an increase in phyllosphere bacterial abundance. Typical phyllosphere-associated taxa such as Sphingomonas, Kineococcus, Friedmaniella, Microbacterium, and Curtobacterium decreased between three and ten-fold in their abundance under aphid herbivory compared to the control. However, in some of the aphid-treated mesocosms, we observed a strong increase in sequence reads affiliated with the genus Buchnera. Buchnera has been described as endosymbiont of aphids (Douglas 1998) and its survival or growth outside the host has not been reported. Consequently, the high abundances of Buchnera are an indicator of the intense aphid infestation, but do not point to a competition-driven functional change in the phyllosphere bacterial community. Insignificant correlations between the amount of total canopy-derived DOC and most of the phyllosphere taxa suggested only a weak response and no stimulation of microbial growth by additional C availability. Notably, 5% of the bacterial taxa were even negatively correlated to the canopy-derived DOC. Among those we identified members of typical phyllosphere-associated families such as Hymenobacteraceae, Microbacteriaceae, Sphingomonadaceae, and Sphingobacteriaceae suggesting a lower tolerance to elevated DOC concentrations.
In fact, exceptionally high local sugar concentrations on the leaf surfaces themselves may have inhibited phyllosphere bacterial growth due to osmotically stressful conditions (Lievens et al. 2015). In a range of up to 12 μg sugar per g moist leaf, Mercier and Lindow (2000) reported a positive relationship between sugar availability on leaf surfaces and the extent of bacterial colonization. However, this is several orders of magnitude lower than the honeydew-derived sugar concentrations available to the leaf microbiota in our experiment (4 to 97.9 mg g−1 dw leaf). This is likely due to the fact that there may be a threshold value and a respective humped-shaped relationship between honeydew-concentration and bacterial growth. In addition, the comparison of electron microscope images of secretion waxes by woolly oak aphids made by others (Ammar et al. 2013) with our images, revealed a clear conformity of wax shapes. These organic molecular structures represent excess wax excretions that densely covered the aphid-infested leaves hence altering the phyllosphere microhabitat structure and making it less amenable to microbial colonisation. Consequently, aphid infestation might initially have triggered bacterial growth by providing honeydew as an additional C source, but this effect diminished as infestation progressed and inhibitory effects of sugar concentration and wax accumulation may have started to dominate.
Michalzik (2011) reported hexose-C proportions in DOC of up to 40–50% entering the topsoil after heavy aphid (Cinara pilicornis) infestation of ten year old Norway spruce. Assuming hexose-C proportions of 40% of total DOC in throughfall, hexose-C amounts of 2–21 and 9–42 kg C * ha−1 * 5d−1 for moderately and heavily infested treatments, respectively, were calculated. Such large amounts of low molecular weight organic substances are very likely to induce responses in the soil. Hence, we suggested that honeydew would induce fast mineralization rates. This effect appeared to be mainly restricted to the organic layer, since the signal of high C-input via honeydew did not translate into organic layer leachates and soil solutions, partly confirming our second hypothesis. The continuous provision of large amounts of C-rich substrates may have fuelled soil microbial activity and might have alleviated microbial C limitation at the aphid treatments. In general, decaying leaf litter is a huge source of dissolved OM, however, as a net result, the readily available C pool more and more depleted with time due to expected accelerated turnover rates and a simultaneous equivalent release of OC from the more stable C pool. We speculate that the frequent honeydew input induced a recurring positive priming effect, stimulating the mineralization of water-extractable fractions of the soil OM pool (Dighton 1978; Dormaar 1990; Michalzik and Stadler 2005). This mechanism was corroborated by an incubation study over 200 days conducted by Zhou et al. (2021) comparing episodic and frequent additions of glucose to a grassland soil. After frequent glucose amendment, a higher proportion of slow-growing, oligotrophic microbes, like saprotrophic fungi that are more capable to mine for N and P in plant litter, were observed. It is possible that frequent honeydew addition especially to the organic layer of our study also increased the fungal abundance in the microbial community, similar to what was found in the topsoil under lime trees (Dighton 1978). In this context, significantly higher bulk δ13C values of the Oi layer of aphid treatments compared to the labelled ones (Fig. 2b) supply evidence for a partial uptake of honeydew-derived C by microbes.
The above-mentioned recurring acceleration of mineralization due to an oversupply of one energy source (honeydew) seemed to have also changed the metabolite composition within all soil layers in the aphid treatment groups. Favouring of specific metabolic processes could have induced the difference in the metabolite profiles. However, the difference seemed to be mainly caused by an increase in the abundance of metabolites and not by the appearance of novel compounds, which in turn, only partially contribute to answer our third hypothesis. Neither an impact of aphid infestation on the chemical richness (number of metabolites produced by soil organisms or exuded by plants) nor on the chemical diversity (Shannon diversity of metabolites) was detected. Hence, none of the metabolites in the chemical profiles was induced or repressed by the aphid attack.
Indirect effects on belowground biochemical properties
In the present study, we assign two possible indirect mechanisms of herbivory to the observed reduced incorporation of freshly assimilated 13C in beech root tissue (Fig. 2a): The first possible indirect mechanism assumes that plants compensate C-losses by diminishing the allocation rate of freshly assimilated resources from shoots to roots. In this context, Smith and Schowalter (2001) reported a translocation of carbohydrates from roots to shoots which led to depleted C-root reserves after severe aphid infestation of Douglas fir seedlings. Additionally, it is also likely that the upwards transfer of C and amino acid with the phloem sap from root to shoot tissue was accelerated by aphids (Girousse et al. 2005) and/or the translocation of freshly assimilated resources down to roots and rhizosphere (Vestergård et al. 2004) was hampered by the aphids itself.
After 2.5 month of aphid infestation, the second indirect mechanism points to a reduction of C-resource allocation rates to roots and rhizodeposits. Between 4 and 27% of the plant’s photosynthetic capacity can be lost due to sap-feeding activity (Dungan et al. 2007; Smith and Schowalter 2001). Accordingly, reductions in the supply of recent photosynthates to belowground plant parts might have cut the exudation of C via roots (Kim et al. 2016; Smith and Schowalter 2001; Vestergård et al. 2004). Similarly, this plant response to severe herbivory infestation may also cause reduced mycorrhizal colonization (Gange et al. 2002; Gehring and Bennett 2009). As a compensation mechanism to the reduced availability of microbial C resources, an acceleration of soil OM decomposition, especially by saprotrophic and/or mycorrhizal fungi in the rooting zone, could have promoted the depletion of the water extractable OC pool.
During the infestation period, the rhizobacterial community and richness may have adapted to the above-mentioned reduced C resource availability (Kim et al. 2016; Liu et al. 2020; Vestergård et al. 2004). In our study, the aphid infestation caused a decrease of rhizosphere bacterial diversity and a community shift, whereas estimated total bacterial abundances were not affected, thus, only partially confirming our fourth hypothesis. Kim et al. (2016) observed a promotion of Paenibacillus sp. growth rate in the rhizosphere of pepper plants by a modulation of rhizodeposition due to aphid infestation. Response patterns in our study were less pronounced and rather reflected the loss of low-abundant taxa upon aphid infestation without clear evidence of a stimulation of particular bacterial taxa in response. Furthermore, the metabolomes of the rhizosphere extracts clearly separated from the control and labelled only treatments assuming changed rhizomicrobial metabolization processes. Consequently, potential changes in rhizosphere microbial functions or plant-microbe feedbacks in response to aphid infestation remain unclear.
Conclusions
Our results revealed distinct direct and indirect effects on plant-insect-microbiome interactions leading to marked alterations in C dynamics upon aphid infestation. This integrative approach provides a stepping stone towards a better understanding of the significance of herbivory-induced alterations in microbial dynamics and element cycles for ecosystem processes. Disentangling and quantifying the relevance of the detected direct and indirect effects of these complex plant-insect-microbiome-interactions on ecosystem processes is crucial. However, to further explain the observed reductions in soil water-extractable OC as well as the impact of relevant groups of (micro-)organisms on the OM and nutrient cycling, a broader community (including saprotrophic and mycorrhizal fungi) and physiological perspective is mandatory. Specific responses of fungal and bacterial phyllosphere communities, such as honeydew C uptake and biomass growth, could be elucidated by additional 13C-determination in characteristic biomarkers. In addition, further studies are required to quantify the extent of priming effects induced by frequent insect-mediated C inputs and to transform insights on plant-insect-microbe-soil systems obtained from mechanistic laboratory studies to the field scale.
Data availability
Sequence data generated in this study have been submitted to the European Nucleotide Archive (ENA) under the study accession number PRJEB46752, sample accession numbers SAMEA9456873 – SAMEA9456904.
Code availability
not available.
References
Ammar E-D, Alessandro RT, Hall DG (2013) Ultrastructural and chemical studies on waxy secretions and wax-producing structures on the integument of the woolly oak aphid Stegophylla brevirostris Quednau (Hemiptera: Aphididae). J Microsc Ultrastruct 1:43–50. https://doi.org/10.1016/j.jmau.2013.05.001
Bardgett RD, Wardle DA (2010) Aboveground-belowground linkages: biotic interactions, ecosystem processes, and global change. Oxford University Press, Oxford
Beggs JR, Karl BJ, Wardle DA, Bonner KI (2005) Soluble carbon production by honeydew scale insects in a New Zealand beech forest. N Z J Ecol 29:105–115
Chelius MK, Triplett EW (2001) The diversity of archaea and bacteria in association with the roots of Zea mays L. Microb Ecol 41:252–263. https://doi.org/10.1007/s002480000087
Chong J, Wishart DS, Xia J (2019) Using MetaboAnalyst 4.0 for comprehensive and integrative metabolomics data analysis. Curr Protoc Bioinformatics 68:e86. https://doi.org/10.1002/cpbi.86
Choudhury D (1985) Aphid honeydew : a re-appraisal of Owen and Wiegert ' s hypothesis. OIKOS 45:287–290
Delory BM, Schempp H, Spachmann SM, Störzer L, van Dam NM, Temperton VM, Weinhold A (2021) Soil chemical legacies trigger species-specific and context-dependent root responses in later arriving plants. Plant Cell Environ 44:1215–1230. https://doi.org/10.1111/pce.13999
Dighton J (1978) Effects of synthetic lime aphid honeydew on populations of soil organisms. Soil Biol Biochem 10:369–376. https://doi.org/10.1016/0038-0717(78)90060-3
Dormaar JF (1990) Effect of active roots on the decomposition of soil organic materials. Biol Fertil Soils 10:121–126. https://doi.org/10.1007/bf00336247
Douglas AE (1998) Nutritional interactions in insect-microbial symbioses: aphids and their symbiotic bacteria Buchnera. Annu Rev Entomol 43:17–37. https://doi.org/10.1146/annurev.ento.43.1.17
Douglas AE (2006) Phloem-sap feeding by animals: problems and solutions. J Exp Bot 57:747–754. https://doi.org/10.1093/jxb/erj067
Dungan RJ, Turnbull MH, Kelly D (2007) The carbon costs for host trees of a phloem-feeding herbivore. J Ecol 95:603–613. https://doi.org/10.1111/j.1365-2745.2007.01243.x
Fischer MK, Volkl W, Hoffmann KH (2005) Honeydew production and honeydew sugar composition of polyphagous black bean aphid, Aphis fabae (Hemiptera : Aphididae) on various host plants and implications for ant-attendance. Eur J Entomol 102: 155-1160. Doi: https://doi.org/10.14411/eje.2005.025
Gange AC, Bower E, Brown VK (2002) Differential effects of insect herbivory on arbuscular mycorrhizal colonization. Oecologia 131:103–112. https://doi.org/10.1007/s00442-001-0863-7
Gehring C, Bennett A (2009) Mycorrhizal fungal–plant–insect interactions: the importance of a community approach*. Environ Entomol 38:93–102. https://doi.org/10.1603/022.038.0111
Girousse C, Moulia B, Silk W, Bonnemain J-L (2005) Aphid infestation causes different changes in carbon and nitrogen allocation in alfalfa stems as well as different inhibitions of longitudinal and radial expansion. Plant Physiol 137:1474–1484. https://doi.org/10.1104/pp.104.057430
Gregorich EG, Beare MH, Stoklas U, St-Georges P (2003) Biodegradability of soluble organic matter in maize-cropped soils. Geoderma 113:237–252. https://doi.org/10.1016/S0016-7061(02)00363-4
Hammer O, Harper D, Ryan P (2001) PAST: paleontological statistics software package for education and data analysis. Palaeontol Electron 4:1–9
Herrmann M, Hadrich A, Küsel K (2012) Predominance of thaumarchaeal ammonia oxidizer abundance and transcriptional activity in an acidic fen. Environ Microbiol 14:3013–3025. https://doi.org/10.1111/j.1462-2920.2012.02882.x
Herrmann M, Geesink P, Richter R, Küsel K (2021) Canopy position has a stronger effect than tree species identity on Phyllosphere bacterial diversity in a floodplain hardwood Forest. Microb Ecol 81:157–168. https://doi.org/10.1007/s00248-020-01565-y
Holland JN, Cheng W, Da C (1996) Herbivore-induced changes in plant carbon allocation: assessment of below-ground C fluxes using carbon-14. Oecologia 107:87–94. https://doi.org/10.1007/BF00582238
Huber W, Carey VJ, Gentleman R, Anders S, Carlson M, Carvalho BS, Bravo HC, Davis S, Gatto L, Girke T, Gottardo R, Hahne F, Hansen KD, Irizarry RA, Lawrence M, Love MI, MacDonald J, Obenchain V, Oleś AK et al (2015) Orchestrating high-throughput genomic analysis with Bioconductor. Nat Methods 12:115–121. https://doi.org/10.1038/nmeth.3252
Jandl R, Sollins P (1997) Water-extractable soil carbon in relation to the belowground carbon cycle. Biol Fertil Soils 25:196–201. https://doi.org/10.1007/s003740050303
Kim B, Song GC, Ryu CM (2016) Root exudation by aphid leaf infestation recruits root-associated Paenibacillus spp. to Lead plant insect susceptibility. J Microbiol Biotechnol 26:549–557. https://doi.org/10.4014/jmb.1511.11058
Kinkel LL (1997) Microbial population dynamics on leaves. Annu Rev Phytopathol 35:327–347. https://doi.org/10.1146/annurev.phyto.35.1.327
Klindworth A, Pruesse E, Schweer T, Peplies J, Quast C, Horn M, Glockner FO (2013) Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res 41. https://doi.org/10.1093/nar/gks808
Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD (2013) Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol 79:5112–5120. https://doi.org/10.1128/aem.01043-13
Krüger M, Potthast K, Michalzik B, Tischer A, Küsel K, Deckner FFK, Herrmann M (2021) Drought and rewetting events enhance nitrate leaching and seepage-mediated translocation of microbes from beech forest soils. Soil Biol Biochem 154:108153. https://doi.org/10.1016/j.soilbio.2021.108153
Kuhl C, Tautenhahn R, Böttcher C, Larson TR, Neumann S (2012) CAMERA: an integrated strategy for compound spectra extraction and annotation of liquid chromatography/mass spectrometry data sets. Anal Chem 84:283–289. https://doi.org/10.1021/ac202450g
Küsel K, Totsche KU, Trumbore SE, Lehmann R, Steinhäuser C, Herrmann M (2016) How deep can surface signals be traced in the critical zone? Merging biodiversity with biogeochemistry research in a central German Muschelkalk landscape. Front Earth Sci 4:1–18
Landsman-Gerjoi M, Perdrial JN, Lancellotti B, Seybold E, Schroth AW, Adair C, Wymore A (2020) Measuring the influence of environmental conditions on dissolved organic matter biodegradability and optical properties: a combined field and laboratory study. Biogeochemistry 149:37–52. https://doi.org/10.1007/s10533-020-00664-9
Lievens B, Hallsworth JE, Pozo MI, Belgacem ZB, Stevenson A, Willems KA, Jacquemyn H (2015) Microbiology of sugar-rich environments: diversity, ecology and system constraints. Environ Microbiol 17:278–298. https://doi.org/10.1111/1462-2920.12570
Liu QP, Li SL, Ding W (2020) Aphid-induced tobacco resistance againstRalstonia solanacearumis associated with changes in the salicylic acid level and rhizospheric microbial community. Eur J Plant Pathol 157:465–483. https://doi.org/10.1007/s10658-020-02005-w
Loy A, Lehner A, Lee N, Adamczyk J, Meier H, Ernst J, Schleifer KH, Wagner M (2002) Oligonucleotide microarray for 16S rRNA gene-based detection of all recognized lineages of sulfate-reducing prokaryotes in the environment. Appl Environ Microbiol 68:5064–5081. https://doi.org/10.1128/aem.68.10.5064-5081.2002
Mercier J, Lindow SE (2000) Role of leaf surface sugars in colonization of plants by bacterial epiphytes. Appl Environ Microbiol 66:369–374. https://doi.org/10.1128/aem.66.1.369-374.2000
Michalzik B (2011) Insects, infestations, and nutrient fluxes. In: Levia DF, Carlyle-Moses DE, Tanaka T (eds) Forest hydrology and biogeochemistry: synthesis of Past research and future directions. Springer, Berlin
Michalzik B, Stadler B (2005) Importance of canopy herbivores to dissolved and particulate organic matter fluxes to the forest floor. Geoderma 127:227–236. https://doi.org/10.1016/j.geoderma.2004.12.006
Michalzik B, Levia DF, Bischoff S, Näthe K, Richter S (2016) Effects of aphid infestation on the biogeochemistry of the water routed through European beech (Fagus sylvatica L.) saplings. Biogeochemistry:1–18. https://doi.org/10.1007/s10533-016-0228-2
Moir ML, Renton M, Hoffmann BD, Leng MC, Lach L (2018) Development and testing of a standardized method to estimate honeydew production. PLoS One 13. https://doi.org/10.1371/journal.pone.0201845
Mühlenberg E, Stadler B (2005) Effects of altitude on aphid-mediated processes in the canopy of Norway spruce. Agric For Entomol 7:133–143. https://doi.org/10.1111/j.1461-9555.2005.00253.x
Nercessian O, Fouquet Y, Pierre C, Prieur D, Jeanthon C (2005) Diversity of Bacteria and Archaea associated with a carbonate-rich metalliferous sediment sample from the rainbow vent field on the mid-Atlantic ridge. Environ Microbiol 7:698–714. https://doi.org/10.1111/j.1462-2920.2005.00744.x
Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O'Hara RB, Simpson GL, Solymos P, Stevens MHH, Wagner H (2015) Vegan: community ecology package. R package vegan, vers 2.2–1
Pathan AK, Bond J, Gaskin RE (2008) Sample preparation for scanning electron microscopy of plant surfaces—horses for courses. Micron 39:1049–1061. https://doi.org/10.1016/j.micron.2008.05.006
Pinheiro J, Bates D, DebRoy S, Sarkar D (2013) Nlme: linear and nonlinear mixed effects models. R core team
Potthast K, Meyer S, Tischer A, Gleixner G, Sieburg A, Frosch T, Michalzik B (2021) Grasshopper herbivory immediately affects element cycling but not export rates in an N-limited grassland system. Ecosphere 12:e03449. https://doi.org/10.1002/ecs2.3449
Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO (2013) The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41:D590–D596. https://doi.org/10.1093/nar/gks1219
R core team (2019) R: a language and environment for statistical computing R Foundation for statistical computing. R core team, Vienna
Ristok C, Poeschl Y, Dudenhöffer J-H, Ebeling A, Eisenhauer N, Vergara F, Wagg C, van Dam NM, Weinhold A (2019) Plant species richness elicits changes in the metabolome of grassland species via soil biotic legacy. J Ecol 107:2240–2254. https://doi.org/10.1111/1365-2745.13185
Rughöft S, Herrmann M, Lazar CS, Cesarz S, Levick SR, Trumbore SE, Küsel K (2016) Community composition and abundance of bacterial, archaeal and nitrifying populations in savanna soils on contrasting bedrock material in Kruger National Park. South Afr Front Microbiol 7. https://doi.org/10.3389/fmicb.2016.01638
Schielzeth H, Forstmeier W (2009) Conclusions beyond support: overconfident estimates in mixed models. Behav Ecol 20:416–420. https://doi.org/10.1093/beheco/arn145
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF (2009) Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541. https://doi.org/10.1128/aem.01541-09
Schowalter TD (2016) Herbivory. Academic Press Ltd-Elsevier Science Ltd, London
Smith JP, Schowalter TD (2001) Aphid-induced reduction of shoot and root growth in Douglas-fir seedlings. Ecol Entomol 26:411–416. https://doi.org/10.1046/j.1365-2311.2001.00335.x
Stadler B, Müller T (2000) Effects of aphids and moth caterpillars on epiphytic microorganisms in canopies of forest trees. Can J For Res-Rev Can Rech Forestiere 30:631–638. https://doi.org/10.1139/cjfr-30-4-631
Stadler B, Solinger S, Michalzik B (2001) Insect herbivores and the nutrient flow from the canopy to the soil in coniferous and deciduous forests. Oecologia 126:104–113. https://doi.org/10.1007/s004420000514
Stoddard S, Smith B, Hein R, Roller B, Schmidt T (2014) Stoddard SF, Smith BJ, Hein R, Roller BRK, Schmidt TM.. rrnDB: improved tools for interpreting rRNA gene abundance in bacteria and archaea and a new foundation for future development. Nucleic Acids Res 43:D593–D598. https://doi.org/10.1093/nar/gku1201
Tautenhahn R, Böttcher C, Neumann S (2008) Highly sensitive feature detection for high resolution LC/MS. BMC Bioinformatics 9:504. https://doi.org/10.1186/1471-2105-9-504
Tischer A, Sehl L, Meyer U-N, Kleinebecker T, Klaus V, Hamer U (2019) Land-use intensity shapes kinetics of extracellular enzymes in rhizosphere soil of agricultural grassland plant species. Plant Soil 437:215–239. https://doi.org/10.1007/s11104-019-03970-w
van Dam NM, Bouwmeester HJ (2016) Metabolomics in the rhizosphere: tapping into belowground chemical communication. Trends Plant Sci 21:256–265. https://doi.org/10.1016/j.tplants.2016.01.008
van Dam NM, Heil M (2011) Multitrophic interactions below and above ground: en route to the next level. J Ecol 99:77–88. https://doi.org/10.1111/j.1365-2745.2010.01761.x
Verhoeven KJF, Simonsen KL, McIntyre LM (2005) Implementing false discovery rate control: increasing your power. Oikos 108:643–647. https://doi.org/10.1111/j.0030-1299.2005.13727.xM4-Citavi
Vestergård M, Bjørnlund L, Christensen S (2004) Aphid effects on rhizosphere microorganisms and microfauna depend more on barley growth phase than on soil fertilization. Oecologia 141:84–93. https://doi.org/10.1007/s00442-004-1651-y
Yang JW, Yi H-S, Kim H, Lee B, Lee S, Ghim S-Y, Ryu C-M (2011) Whitefly infestation of pepper plants elicits defence responses against bacterial pathogens in leaves and roots and changes the below-ground microflora. J Ecol 99:46–56. https://doi.org/10.1111/j.1365-2745.2010.01756.x
Zhou J, Wen Y, Shi LL, Marshall MR, Kuzyakov Y, Blagodatskaya E, Zang HD (2021) Strong priming of soil organic matter induced by frequent input of labile carbon. Soil Biol Biochem 152:11. https://doi.org/10.1016/j.soilbio.2020.108069
Zoebelein G (1954) Versuche zur Feststellung des Honigtauertrages von Fichtenbeständen mit Hilfe von Waldameisen. Z Angew Entomol 36:358–362. https://doi.org/10.1111/j.1439-0418.1954.tb00764.x
Zsolnay Á (2003) Dissolved organic matter: artefacts, definitions, and functions. Geoderma 113:187–209. https://doi.org/10.1016/S0016-7061(02)00361-0
Acknowledgements
This study is part of the Collaborative Research Centre AquaDiva of the Friedrich Schiller University Jena, funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) – SFB 1076 –Project Number 218627073. We thank Christine Hess and Maria Fabisch for scientific coordination. AW and NMvD were supported by the German Centre for Integrative Biodiversity Research (iDiv) Halle-Jena-Leipzig funded by the German Research Foundation (DFG–FZT 118, 202548816). We acknowledge Sanjay Lama for assistance in the lab. Climate chambers to conduct experiments under controlled temperature and light conditions, and the instrumentation to perform Illumina MiSeq amplicon sequencing were financially supported by the Thüringer Ministerium für Wirtschaft, Wissenschaft und Digitale Gesellschaft (TMWWDG; project B 715-09075, and project 2016 FGI 0024 “BIODIV”, respectively). For the customized production of the mesocosms we thank Martin Stephan and his colleagues of the workshop of the Otto-Schott-Institute for Materials Research, Friedrich Schiller University Jena. Heidrun Garlipp (Chair of Material Science, Friedrich Schiller University Jena) is thanked for the SEM imaging, which is funded by the Deutsche Forschungsgemeinschaft DFG (grant reference INST 275/241-1 FUGG) and by the Thüringer Ministerium für Bildung, Wissenschaft und Kultur TMBWK (grant reference 62-4264 925/1/10/1/01). The authors gratefully acknowledge our technician Bernd Ruppe for taking care of the plant growth in the mesocosms. We thank Heike Geilmann of the Max Planck Institute for Biogeochemistry in Jena for 13C isotope analyses. Bastian Polst, Linda Gorniak and Stefan Riedel are acknowledged for support of DNA extraction, amplicon library preparation, and Illumina MiSeq sequencing. We thank the anonymous reviewers for their valuable suggestions and comments on the manuscript.
Funding
Open Access funding enabled and organized by Projekt DEAL. This study is part of the Collaborative Research Centre AquaDiva of the Friedrich Schiller University Jena, funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) – SFB 1076 –Project Number 218627073. AW and NMvD were supported by the German Centre for Integrative Biodiversity Research (iDiv) Halle-Jena-Leipzig funded by the German Research Foundation (DFG–FZT 118, 202548816). Climate chambers to conduct experiments under controlled temperature and light conditions, and the instrumentation to perform Illumina MiSeq amplicon sequencing were financially supported by the Thüringer Ministerium für Wirtschaft, Wissenschaft und Digitale Gesellschaft (TMWWDG; project B 715–09075, and project 2016 FGI 0024 “BIODIV”, respectively). Heidrun Garlipp (Chair of Material Science, Friedrich Schiller University Jena) is thanked for the SEM imaging, which is funded by the Deutsche Forschungsgemeinschaft DFG (grant reference INST 275/241–1 FUGG) and by the Thüringer Ministerium für Bildung, Wissenschaft und Kultur TMBWK (grant reference 62–4264 925/1/10/1/01).
Author information
Authors and Affiliations
Contributions
KP and AT planned the experimental design and sampling strategy and performed the experiment, realizing BM’s original research idea. KP and AT did the chemical analyses of ecosystem solutions and bulk samples and its data processing. MH and KK did the microbial analyses including molecular work and sequence data processing. AW and NMvD performed the metabolomics analyses and its data processing. KP, BM, AT, MH wrote the manuscript with contributions from all other authors.
Corresponding author
Ethics declarations
Conflicts of interest/ competing interests
no conflicts.
Additional information
Responsible Editor: Katharina Maria Keiblinger.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Potthast, K., Tischer, A., Herrmann, M. et al. Woolly beech aphid infestation reduces soil organic carbon availability and alters phyllosphere and rhizosphere bacterial microbiomes. Plant Soil 473, 639–657 (2022). https://doi.org/10.1007/s11104-022-05317-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11104-022-05317-4