Abstract
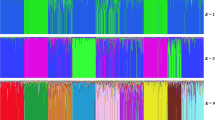
The Nguni cattle breed has distinct populations that are adapted to the different ecological zones of Southern Africa. This study was carried out to assess genetic diversity and establish the relationships among South African (SA), Mozambican (Landim), and Swazi Nguni cattle populations, using 25 microsatellite markers. Genotypic data were generated from deoxyribonucleic acid (DNA) samples of 90 unrelated individuals of the three cattle populations, collected from government conservations and stud herds. DNA profiles of five local beef breeds were used as the reference populations. Most of the 25 microsatellite markers were highly polymorphic across the studied populations, with an overall polymorphic information content (PIC) mean of 0.676. Genetic diversity within populations was high with expected heterozygosity varying from 0.705 ± 0.024 (Landim) to 0.748 ± 0.021 (SA Nguni) and mean number of alleles being highest in the SA Nguni (7.52 ± 0.42). Average observed heterozygosity (0.597 ± 0.046) compared to the expected heterozygosity (0.719 ± 0.022) was lowest for the Swazi Nguni, which also had a high number of Hardy-Weinberg Equilibrium (HWE) deviated loci (13), confirming the relatively high level of inbreeding (0.158 ± 0.058) in that population. Analysis of molecular variance revealed only 9.61% of the total variation between the populations and 90.39% within populations. A short genetic distance (0.299) was observed between Landim and Swazi Nguni, with the SA Nguni (> 0.500) being the most genetically distant population. The distant relationship between SA Nguni and the other two Nguni cattle populations was further confirmed by a principal coordinates analysis. The three Nguni populations clustered independently from each other, despite some evidence of admixture. Therefore, it can be concluded that SA Nguni, Landim, and Swazi Nguni populations in Southern Africa exhibit high levels of genetic diversity and are genetically distant; with the two latter populations being less genetically apart. These results present useful information for the development of strategies for regional management of animal genetic resources, through conservation and sustainable utilisation.




Similar content being viewed by others
References
Acosta, A.C., Uffo, O., Sanz, A., Ronda, R., Osta, R., Rodellar, C., Martin-Burriel, I., Zaragoza, P., 2013. Genetic diversity and differentiation of five Cuban cattle breeds using 30 microsatellite loci. Journal of Animal Breeding and Genetics, 130 (1), 79–86.
Bessa, I., Pinheiro, I., Matola, M., Dzama, K., Rocha A. and Alexandrino P., 2009. Genetic diversity and relationships among indigenous Mozambican cattle breeds. South African Journal of Animal Science, 39 (1), 61–75.
Bester, J., Matjuda, L.E., Rust, J.M. and Fourie, H.J., 2003. The Nguni: A Case Study. In Vilakati et al. (eds) 2003. Community-based management of animal genetic resources. Rome FAO.
Botstein, D., White, R.L., Skolnick, M. and Davis, R.W., 1980. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. American Journal of Human Genetics, 32, 314–331.
Dadi, H., Tibbo, M., Takahashi, Y., Nomura, K., Hanada, H. and Amano, T., 2008. Microsatellite analysis reveals high genetic diversity but low genetic structure in Ethiopian indigenous cattle populations. Animal Genetics, 39, 425–431.
Earl, D.A. and vonHoldt, B.M., 2012. Structure Harvester: A website and program for visualizing STRUCTURE output and implementing the Evanno’s method. Conservation Genetics Resources, 4, 359–361.
Evanno, G., Regnaut, S. and Goudet, J., 2005. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Molecular Ecology, 14, 2611–2620.
Excoffier, L., Laval, G. and Schneider, S., 2005. Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evolutionary Bioinformatics Online, 1, 47–50.
FAO, 2007. The state of the world’s animal genetic resources for food and agriculture. FAO, Rome.
FAO, 2011. Molecular genetic characterization of animal genetic resources. FAO Animal Production and Health Guidelines. No.9. In: Commission on Genetic Resources for Food and Agriculture Assessments. FAO, Rome.
FAO, 2012. Status and trends of Animal Genetics Resources. Item 4.2 of the Provisional Agenda. Fourteenth Regular Session. Rome, 15–19 April.
Farmer’s weekly, 2013. Saving the Swazi Nguni. 20–27 December.
Gautier, M., Faraut, T., Moazami-Goudarzi, K., Navratil, V., Foglio, M., Grohs, C., Boland, A., Garnier, J., Boichard, D., Lathrop, G.M., Gut, I.G. and Eggen, A., 2007. Genetic and Haplotypic Structure in European and African cattle breeds. Genetics Society of America, 177, 1059–1070.
Gororo, E., Makuza, S.M., Chatiza, F.P., Chidzwondo, F. and Sanyika, T.W., 2018. Genetic diversity in Zimbabwean Sanga cattle breeds using microsatellite markers. South African Journal of Animal Science, 48 (1), 128–141.
Groeneveld, L.F., Lenstra, J.A., Eding, H., Toro, M.A., Scherf, B., Pilling, D., Negrini, R., Finlay, E.K., Jianlin, H., Groeneveld, E. and Weigend, S., 2010. Genetic diversity in farm animals – a review. Animal Genetics, 41, 6–31.
Hlophe, R.S., 2011. Genetic variation between and within six selected South African sheep breeds using random amplified polymorphic DNA and protein markers. MSc dissertation. University of Zululand, South Africa.
Horsburgh, K.A., Prost, S., Gosling, A., Stanton, J.A., Rand, C. and Matisoo-Smith, E.A., 2013. The Genetic Diversity of the Nguni Breed of African Cattle (Bos spp.): Complete Mitochondrial Genomes of Haplogroup T1. PloS ONE 8(8), e71956. Retrieved October 20, 2017, from https://doi.org/10.1371/journal.pone.0071956
Kunene, N.W., Nesamvuni, E.A. and Fossey, A., 2007. Characterization of Zulu (Nguni) sheep using linear body measurements and some environmental factors affecting these measurements. South African Journal of Animal Science, 37 (1), 11–20.
Madilindi, M.A., Banga, C.B., Bhebhe, E., Sanarana, Y.P., Nxumalo, K.S., Taela, M.G. and Mapholi, N.O., 2019. Genetic differentiation and population structure of four Mozambican indigenous cattle populations. Livestock Research for Rural Development, 31 (47). Retrieved April 4, 2019, from http://www.lrrd.org/lrrd31/4/matom31047.html
Mapholi, N.O., Marufu, M.C., Maiwashe, A., Banga, C.B. Muchenje, V., MacNeil, M.D., Chimonyo, M. and Dzama, K., 2014. Towards a genomics approach to tick (Acari: Ixodidae) control in cattle: A review. Ticks Tick-borne Diseases, 5(5), 475–483.
Mapholi, N.O., Maiwashe, A., Matika, O., Riggio, V., Bishop, S.C., MacNeil, M.D., Banga, C., Taylor J.F. and Dzama, K., 2016. Genome-wide association study of tick resistance in South African Nguni cattle. Ticks Tick-Borne Diseases, 7, 487–497.
Marufu, M.C., Qokweni, L., Chimonyo, M. and Dzama, K., 2011. Relationships between tick counts and coat characteristics in Nguni and Bonsmara cattle reared on semiarid rangelands in South Africa. Ticks Tick-Borne Diseases, 2, 172–177.
Mwai, O., Hanotte, O., Kwon, Y.J. and Cho, S., 2015. African indigenous cattle: unique genetic resources in a rapidly changing world. Asian-Australasian Journal Animal Science, 28, 911–21.
Ndiaye, N.P., Sow, A., Dayo, G.K., Ndiaye, S., Sawadogo, G.J. and Sembène, M., 2015. Genetic diversity and phylogenetic relationships in local cattle breeds of Senegal based on autosomal microsatellite markers. Veterinary World Journal, 8 (8), 994–1005.
Ndumu, D.B., Baumung, R., Hanotte, O., Wurzinger, M., Okeyo, M.A., Jianlin, H., Kibogo, H. and Solkner, J., 2008. Genetic and morphological characterization of the Ankole Longhorn cattle in the African Great Lakes region. Genetic Selection Evolution, 40, 467–490.
Nei, M. 1987. Molecular evolutionary genetics. New York, Columbia University Press.
Ngono-Ema, P.J., Manjeli, Y., Meutchieyié, F., Keambou, C., Wanjala, B., Desta A.F., Ommeh, S., Skilton, R. and Djikeng, A., 2014. Genetic diversity of four Cameroonian indigenous cattle using microsatellite markers. Journal of Livestock Sciences, 5, 9–17.
Nyamushamba, G., Mapiye, C., Tada, O., Halimani, T. and Muchenje, V., 2017. Conservation of indigenous cattle genetic resources in Southern Africa’s smallholder areas: Turning threats into opportunities – A review. Asian-Australasian Journal Animal Science, 30, 603–622.
Padilla, J.A., Sansinforiano, E., Parejo, J.C., Rabasco, A., Martínez-Trancón, M., 2009. Inference of admixture in the endangered Blanca Cacereña bovine breed by microsatellite analyses. Livestock Science, 122 (2-3), 314–322.
Park, S.D.E, 2001. Trypanotolerance in West African and the population genetics effects of selection. PhD Thesis. University of Dublin, Dublin, Ireland.
Peakall, R. and Smouse, P.E., 2006. GenAlex 6: genetic analysis in excel. Population genetic software for teaching and research. Molecular Ecology, 6, 288–295.
Perrier, X., and Jacquemoud-Collet, J.P., 2006. DARwin software. Retrieved July 07, 2017, from http://darwin.cirad.fr/
Pham, L.D., Do, D.N., Binh, N.T., Nam, L.Q., Ba, N.V., Thuy, T.T.T., Hoan, T.X., Cuong, V.C., and Kadarmideen, H.N., 2013. Assessment of genetic diversity and population structure of Vietnamese indigenous cattle populations by microsatellites. Livestock Science, 155, 17–22.
Prichard, J.K., Stephens, M. and Donnelly, P., 2000. Inference of population structure using multilocus genotype data. Genetics, 155, 945–959.
Raymond, M. and Rousset, F., 1995. Genepop (Version-1.2) – population-genetics software for exact tests. Journal of Heredity. 86, 248–249.
Rege, J.E.O. and Tawah, C.L., 1999. The state of African cattle genetic resources II. Geographical distribution, characteristics and uses of present-day breeds and strains. Animal Genetic Resources Information, No. 26.
Sambrook, J., Fritsch, E.F. and Maniatis, T., 1989. Molecular cloning: A laboratory manual, volume 3. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y, USA.
Sanarana, Y., Visser, C., Bosman, L., Nephawe, K., Maiwashe, A. and van Marle-Köster, E., 2016. Genetic diversity in South African Nguni cattle ecotypes based on microsatellites markers. Tropical Animal Health and Production, 48 (2), 379–385.
Scholtz, M.M. and Ramsay, K.A., 2007. Experience in establishing a herd book for the local Nguni breed in South Africa. Agriculture, 41, 25–28.
Scholtz, M.M., Gertenbach, W.D. and Hallowell, G.J., 2011. The Nguni breed of cattle: Past, Present and Future. The Nguni cattle Breeder’s Society, Pretoria, South Africa.
Sharma, R., Maitra, A., Singh, K.P. and Tantia, S.M., 2013. Genetic diversity and relationship of cattle populations of East India: distinguishing lesser known cattle populations and established breeds based on STR markers. Springer Plus, 2 (359), 1–10.
Sharma, R., Kishore, A., Mukesh, M., Ahlawat, S., Maitra, A., Pandey, A.K. and Tantia, M.S., 2015. Genetic diversity and relationship of Indian cattle inferred from microsatellite and mitochondrial DNA markers. BioMed Central Genetics, 16 (73), 1–12.
Van de Pypekamp, H.E., 2013. Nguni Cattle: Breed Characteristics and Functional Efficiency. First edition. Media24 Weekly Magazines 40 Heerengracht, Cape Town 8000: Landbouweeklad.
Zwane, A.A., Schnabel, R.D., Hoff, J., Choudhury, A., Makgahlela, M.L., Maiwashe, A., van Marle Köster, E. and Taylor, J.F., 2019. Genome-Wide SNP Discovery in Indigenous Cattle Breeds of South Africa. Frontiers in Genetics. 10, 273. doi: https://doi.org/10.3389/fgene.2019.00273
Acknowledgments
The authors would like to duly acknowledge the collaboration and contribution provided by the Agricultural Research Council, Animal Production, South Africa; Agrarian Research Institute of Mozambique, Directorate of Animal Science, Mozambique; and Ministry of Agriculture, Department of Livestock and Veterinary Services, Swaziland, for the execution of this study.
Funding
This project received financial support from the Technology Innovation Agency, National Research Foundation (Grant Number 103042), University of Venda: Research and Publication Committee grant (SARDF/16/ANS/03/1404) and Southern African Science Service Centre for Climate Change and Adaptive Land Management (SASSCAL) - Council for Scientific and Industrial Research (CSIR) of South Africa.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Statement of animal rights
All the international, regional, national and institutional guidelines for the care and use of animals were followed. Approval of the study was granted by the Animal Ethics Committee (AEC) of the University of Venda, South Africa (SARDF/16/ANS/03/1404).
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Madilindi, M.A., Banga, C.B., Bhebhe, E. et al. Genetic diversity and relationships among three Southern African Nguni cattle populations. Trop Anim Health Prod 52, 753–762 (2020). https://doi.org/10.1007/s11250-019-02066-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11250-019-02066-y