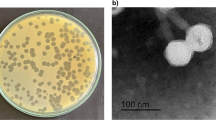
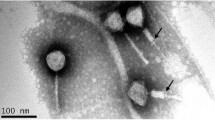
Abstract
Pseudomonas aeruginosa is an opportunistic human pathogen that can lead to nosocomial infections which are in turn life threatening. The increase in antibiotic resistance, at an alarming rate, has resulted in a pressing need for alternative therapeutic approaches such as phage therapy, which hold promise according to several studies. This study featured the isolation and characterization of vB_PaeS_TUMS_P81, a new lytic Pseudomonas phage. The whole-genome sequencing indicated that it has a genome of 73,167 bp containing 93 predicted coding sequences. Genes involved in virulence or lysogeny pathway were nowhere to be found in the genome, so it is potentially safe when it comes to therapeutic applications. Genomic and phylogenetic analysis indicated that vB_PaeS_TUMS_P81 is a member of the genus Litunavirus, belonging to Schitoviridae family. The present study lays the groundwork for further research on treatment of P. aeruginosa infections.









Similar content being viewed by others
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Tenover FC, Nicolau DP, Gill CM (2022) Carbapenemase-producing Pseudomonas aeruginosa -an emerging challenge. Emerg Microbes Infect 11:811–814. https://doi.org/10.1080/22221751.2022.2048972
Yaeger LN, Coles VE, Chan DCK, Burrows LL (2021) How to kill Pseudomonas-emerging therapies for a challenging pathogen. Ann N Y Acad Sci 1496:59–81. https://doi.org/10.1111/nyas.14596
Lister PD, Wolter DJ, Hanson ND (2009) Antibacterial-resistant Pseudomonas aeruginosa: clinical impact and complex regulation of chromosomally encoded resistance mechanisms. Clin Microbiol Rev 22:582–610. https://doi.org/10.1128/cmr.00040-09
Antonelli G, Cappelli L, Cinelli P, Cuffaro R, Manca B, Nicchi S et al (2021) Strategies to tackle antimicrobial resistance: the example of escherichia coli and pseudomonas aeruginosa. Int J Mol Sci 22:4943. https://doi.org/10.3390/ijms22094943
Tacconelli E, Magrini N, Kahlmeter G, Singh N (2017) Global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics. World Health Organization 27:318–327
Tümmler B (2019) Emerging therapies against infections with Pseudomonas aeruginosa. F1000Res 8:1371. https://doi.org/10.12688/f1000research.19509.1
Weinbauer MG (2004) Ecology of prokaryotic viruses. FEMS Microbiol Rev 28:127–181. https://doi.org/10.1016/j.femsre.2003.08.001
Zschach H, Joensen KG, Lindhard B, Lund O, Goderdzishvili M, Chkonia I et al (2015) What can we learn from a metagenomic analysis of a georgian bacteriophage cocktail? Viruses 7:6570–6589. https://doi.org/10.3390/v7122958
Fauconnier A (2019) Phage therapy regulation: from night to dawn. Viruses. https://doi.org/10.3390/v11040352
Pirnay JP, Blasdel BG, Bretaudeau L, Buckling A, Chanishvili N, Clark JR et al (2015) Quality and safety requirements for sustainable phage therapy products. Pharm Res 32:2173–2179. https://doi.org/10.1007/s11095-014-1617-7
Spilker T, Coenye T, Vandamme P, LiPuma JJ (2004) PCR-based assay for differentiation of Pseudomonas aeruginosa from other Pseudomonas species recovered from cystic fibrosis patients. J Clin Microbiol 42:2074–2079. https://doi.org/10.1128/JCM.42.5.2074-2079.2004
Kropinski AM, Mazzocco A, Waddell TE, Lingohr E, Johnson RP (2009) Enumeration of bacteriophages by double agar overlay plaque assay. Methods Mol Biol 501:69–76. https://doi.org/10.1007/978-1-60327-164-6_7
Sillankorva S (2018) Isolation of Bacteriophages for Clinically Relevant Bacteria. Methods Mol Biol 1693:23–30. https://doi.org/10.1007/978-1-4939-7395-8_3
Kropinski AM (2009) Measurement of the rate of attachment of bacteriophage to cells. Methods Mol Biol 501:151–155. https://doi.org/10.1007/978-1-60327-164-6_15
Kropinski AM (2018) Practical advice on the one-step growth curve. In: Martha RJC, Andrew MK, Rob L (eds) Bacteriophages Methods and Protocols. Springer, NY, pp 41–47. https://doi.org/10.1007/978-1-4939-7343-9_3
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. https://doi.org/10.1093/bioinformatics/btu170
Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS et al (2012) SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. https://doi.org/10.1089/cmb.2012.0021
Wick RR, Schultz MB, Zobel J, Holt KE (2015) Bandage: interactive visualization of de novo genome assemblies. Bioinformatics 31:3350–3352. https://doi.org/10.1093/bioinformatics/btv383
Garneau J, Depardieu F, Fortier L-C, Bikard D, Monot M (2017) PhageTerm: a fast and user-friendly software to determine bacteriophage termini and packaging mode using randomly fragmented NGS data. Sci Rep. https://doi.org/10.1038/s41598-017-07910-5
Li H, Durbin R (2010) Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26:589–595. https://doi.org/10.1093/bioinformatics/btp698
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N et al (2009) The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. https://doi.org/10.1093/bioinformatics/btp352
Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA et al (2008) The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9:1–15. https://doi.org/10.1186/1471-2164-9-75
Arndt D, Grant JR, Marcu A, Sajed T, Pon A, Liang Y et al (2016) PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res. https://doi.org/10.1093/nar/gkw387
Moraru C, Varsani A, Kropinski AM (2020) VIRIDIC-A novel tool to calculate the intergenomic similarities of prokaryote-infecting viruses. Viruses 12:1268. https://doi.org/10.3390/v12111268
Lowe TM, Eddy SR (1997) tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 25:955–964. https://doi.org/10.1093/nar/25.5.955
Tynecki P, Guziński A, Kazimierczak J, Jadczuk M, Dastych J, Onisko A (2020) PhageAI - Bacteriophage life cycle recognition with machine learning and natural language processing. bioRxiv.https://doi.org/10.1101/2020.07.11.198606
Jia B, Raphenya AR, Alcock B, Waglechner N, Guo P, Tsang KK et al (2016) CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. https://doi.org/10.1093/nar/gkw1004
Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O et al (2012) Identification of acquired antimicrobial resistance genes. J Antimicrobial Chemotherapy 67:2640–2644. https://doi.org/10.1093/jac/dks261
Gupta SK, Padmanabhan BR, Diene SM, Lopez-Rojas R, Kempf M, Landraud L et al (2014) ARG-ANNOT, a new bioinformatic tool to discover antibiotic resistance genes in bacterial genomes. Antimicrobial Agents Chemotherapy 58:212–220. https://doi.org/10.1128/AAC.01310-13
Chen L, Zheng D, Liu B, Yang J, Jin Q (2016) VFDB 2016: hierarchical and refined dataset for big data analysis—10 years on. Nucleic acids research 44:D694-D7.https://doi.org/10.1093/nar/gkv1239
Carattoli A, Zankari E, García-Fernández A, Voldby Larsen M, Lund O, Villa L et al (2014) In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrobial Agents Chemotherapy 58:3895–3903. https://doi.org/10.1128/AAC.02412-14
Seemann T (2016) ABRicate: mass screening of contigs for antiobiotic resistance genes. Available from: https://github.com/tseemann/abricate.
Adriaenssens E, Brister JR (2017) How to name and classify your phage: an informal guide. Viruses 9:70. https://doi.org/10.3390/v9040070
Wittmann J, Turner D, Millard AD, Mahadevan P, Kropinski AM, Adriaenssens EM (2020) From orphan phage to a proposed new family-the diversity of N4-like viruses. Antibiotics (Basel) 9:663. https://doi.org/10.3390/antibiotics9100663
Tamura K, Stecher G, Kumar S (2021) MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol Biol Evolution 38:3022–3027. https://doi.org/10.1093/molbev/msab120
Stothard P, Wishart DS (2005) Circular genome visualization and exploration using CGView. Bioinformatics 21:537–539. https://doi.org/10.1093/bioinformatics/bti054
Sullivan MJ, Petty NK, Beatson SA (2011) Easyfig: a genome comparison visualizer. Bioinformatics 27:1009–1010. https://doi.org/10.1093/bioinformatics/btr039
Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG et al (2012) Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect 18:268–281. https://doi.org/10.1111/j.1469-0691.2011.03570.x
Gu J, Liu X, Li Y, Han W, Lei L, Yang Y et al (2012) A method for generation phage cocktail with great therapeutic potential. PLoS ONE 7:e31698. https://doi.org/10.1371/journal.pone.0031698
Oechslin F, Piccardi P, Mancini S, Gabard J, Moreillon P, Entenza JM et al (2017) Synergistic interaction between phage therapy and antibiotics clears pseudomonas aeruginosa infection in endocarditis and reduces virulence. J Infect Dis 215:703–712. https://doi.org/10.1093/infdis/jiw632
Ceyssens PJ, Brabban A, Rogge L, Lewis MS, Pickard D, Goulding D et al (2010) Molecular and physiological analysis of three Pseudomonas aeruginosa phages belonging to the “N4-like viruses.” Virology 405:26–30. https://doi.org/10.1016/j.virol.2010.06.011
Choi KH, McPartland J, Kaganman I, Bowman VD, Rothman-Denes LB, Rossmann MG (2008) Insight into DNA and protein transport in double-stranded DNA viruses: the structure of bacteriophage N4. J Mol Biol 378:726–736. https://doi.org/10.1016/j.jmb.2008.02.059
Russell DA (2018) Sequencing, assembling, and finishing complete bacteriophage genomes. Methods Mol Biol 1681:109–125. https://doi.org/10.1007/978-1-4939-7343-9_9
Fang X, Fang Z, Zhao J, Zou Y, Li T, Wang J et al (2012) Draft genome sequence of Pseudomonas aeruginosa strain ATCC 27853. J Bacteriol 194:3755. https://doi.org/10.1128/jb.00690-12
Kwan T, Liu J, Dubow M, Gros P, Pelletier J (2006) Comparative genomic analysis of 18 Pseudomonas aeruginosa bacteriophages. J Bacteriol 188:1184–1187. https://doi.org/10.1128/jb.188.3.1184-1187.2006
Paddison P, Abedon ST, Dressman HK, Gailbreath K, Tracy J, Mosser E et al (1998) The roles of the bacteriophage T4 r genes in lysis inhibition and fine-structure genetics: a new perspective. Genetics 148:1539–1550. https://doi.org/10.1093/genetics/148.4.1539
Acknowledgements
We thank Dr. M. Douraghi, from Tehran University of Medical Sciences, IKHC and Noor Reference Laboratory for providing bacterial strains; Dr. S. Sabouri Shahrbabak for her kind assistance; and Mr. S. Delara for his invaluable help writing the manuscript.
Funding
This work was supported by the National Institute for Medical Research Development (NIMAD), Grant no. 964206.
Author information
Authors and Affiliations
Contributions
Haniyeh Kamyab: Data curation; Formal analysis; Investigation; Methodology; Writing -original draft; Software. Narges Torkashvand: Data curation; Formal analysis; Investigation; Methodology; Software. Ahmad Reza Shahverdi: Supervision, Project administration; Funding acquisition. Mohammad Reza Khoshayand: Supervision; Validation. Mohammad Sharifzadeh: Supervision; Validation. Zargham Sepehrizadeh: Conceptualization, Supervision, Project administration, Funding acquisition, Writing—review and editing.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no known competing financial interests or personal relationships that could have appeared to influence the work reported.
Additional information
Edited by Andrew Millard.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kamyab, H., Torkashvand, N., Shahverdi, A.R. et al. Isolation, characterization, and genomic analysis of vB_PaeS_TUMS_P81, a lytic bacteriophage against Pseudomonas aeruginosa. Virus Genes 59, 132–141 (2023). https://doi.org/10.1007/s11262-022-01954-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11262-022-01954-0