Abstract
Introduction
Fusarium oxysporum has a high affinity for lignin and cellulose-based substrates and is known to grow in a wide range of environments. It is these properties and its ability to produce mycotoxins that have contributed to its pathogenicity in cereal crops that can affect human and animal health when ingested.
Objectives
Identify the mechanisms of mycotoxin production and map the functional output of F. oxysporum under varying growth conditions.
Methods
Liquid and gas-based chromatography coupled with mass spectrometry was used to identify and map the untargeted metabolic pathway of F. oxysporum grown using nitrogen limited and organic/inorganic nitrogen supplemented media.
Results
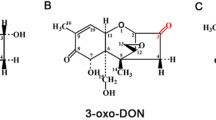
Over 1300 metabolites were identified, relating to 42 metabolic pathways. Of these, 520 metabolites merged at pyruvate (glycolysis), succinate (Krebs cycle) and aspartate-glutamate metabolic pathways. CoA depletion at the growth stage triggered the initiation of fatty acid and branched amino acid degradation. This in turn activated propionyl CoA carnitine acetyltransferase enzymes, resulting in nitrogen preservation (urea, putrescine and organic acids end-products). CoA then transferred into the TCA cycle via previously unreported β-alanine and propionyl CoA metabolic pathways, the latter likely being a novel methylmalonyl-CoA mutase activity for F. oxysporum.
Conclusions
The lower supplementation of inorganic nitrogen compounds (≤ 50 mM) and the elimination of nitrates/organic nitrogen sources resulted in TCA autophagy events that boosted mycotoxin-based metabolism and decreased overall F. oxysporum growth. Such knowledge of functional mycotoxin production can be used to supplement agricultural crops and reduce the risk of mycotoxin contamination in human and animal food supplies.




Similar content being viewed by others
References
Beale, D. J., Karpe, A. V., & Ahmed, W. (2016). Beyond metabolomics: A review of multi-omics-based approaches. In Microbial metabolomics (pp. 289–312). New York: Springer.
Beale, D. J., Pinu, F. R., Kouremenos, K. A., Poojary, M. M., Narayana, V. K., Boughton, B. A., et al. (2018). Review of recent developments in gc–ms approaches to metabolomics-based research. Metabolomics, 14, 152.
Bouras, N., Holtz, M. D., Aboukhaddour, R., & Strelkov, S. E. (2016). Influence of nitrogen sources on growth and mycotoxin production by isolates of pyrenophora tritici-repentis from wheat. The Crop Journal, 4, 119–128.
Brown, M., Wedge, D. C., Goodacre, R., Kell, D. B., Baker, P. N., Kenny, L. C., Mamas, M. A., Neyses, L., & Dunn, W. B. (2011). Automated workflows for accurate mass-based putative metabolite identification in lc/ms-derived metabolomic datasets. Bioinformatics, 27, 1108–1112.
Carroll, A. J., Zhang, P., Whitehead, L., Kaines, S., Tcherkez, G., & Badger, M. R. (2015). Phenometer: A metabolome database search tool using statistical similarity matching of metabolic phenotypes for high-confidence detection of functional links. Frontiers in Bioengineering and Biotechnology, 3, 106.
Chen, J., Sutter, B. M., Shi, L., & Tu, B. P. (2017). Gator1 regulates nitrogenic cataplerotic reactions of the mitochondrial tca cycle. Nature Chemical Biology, 13, 1179.
Christakopoulos, P., Macris, B. J., & Kekos, D. (1989). Direct fermentation of cellulose to ethanol by fusarium oxysporum. Enzyme and Microbial Technology, 11, 236–239.
Dalpé, Y., Trépanier, M., Sahraoui, A. L.-H., Fontaine, J., & Sancholle, M. (2012). 8 lipids of mycorrhizas. In B. Hock (Ed.), Fungal associations (pp. 137–169). Berlin Heidelberg: Springer.
Fiehn, O., Robertson, D., Griffin, J., van der Werf, M., Nikolau, B., Morrison, N., et al. (2007). The metabolomics standards initiative (msi). Metabolomics, 3, 175–178.
Gromski, P. S., Muhamadali, H., Ellis, D. I., Xu, Y., Correa, E., Turner, M. L., & Goodacre, R. (2015). A tutorial review: Metabolomics and partial least squares-discriminant analysis – a marriage of convenience or a shotgun wedding. Analytica Chimica Acta, 879, 10–23.
Gunnaiah, R. (2013). Functional characterization of wheat fusarium head blight resistance qtl (fhb1) based on non-targeted metabolomics and proteomics. Montreal: McGill University.
Haider, S., & Pal, R. (2013). Integrated analysis of transcriptomic and proteomic data. Current Genomics, 14, 91–110.
HiMedia (2012) Rm006: Mycological peptone Culture media bases, HiMedia Laboratories Pvt. Ltd.
Horai, H., Arita, M., Kanaya, S., Nihei, Y., Ikeda, T., Suwa, K., et al. (2010). Massbank: A public repository for sharing mass spectral data for life sciences. Journal of Mass Spectrometry, 45, 703–714.
Jonkers, W., Rodrigues, C. D. A., & Rep, M. (2009). Impaired colonization and infection of tomato roots by the δfrp1 mutant of fusarium oxysporum correlates with reduced cwde gene expression. Molecular Plant-Microbe Interactions, 22, 507–518.
Karine, P., Paul, A., André, G., & Russell, J. T. (2006). Fatty acid composition of lipids from mushrooms belonging to the family boletaceae. Mycological Research, 110, 1179–1183.
Karpe, A. V., Beale, D. J., Godhani, N. B., Morrison, P. D., Harding, I. H., & Palombo, E. A. (2016). Untargeted metabolic profiling of winery-derived biomass waste degradation by aspergillus niger. Journal of Chemical Technology & Biotechnology, 91, 1505–1516.
Karpe, A. V., Beale, D. J., Morrison, P. D., Harding, I. H., Palombo, E. A., & Boden, R. (2015). Untargeted metabolic profiling of vitis vinifera during fungal degradation. FEMS Microbiology Letters, 362, fnv060–fnv060.
Kazan, K., & Gardiner, D. M. (2017). Transcriptomics of cereal–fusarium graminearum interactions: What we have learned so far. Molecular Plant Pathology, 19(3), 764–778.
Kosová, K., Chrpová, J., Šantrůček, J., Hynek, R., Štěrbová, L., Vítámvás, P., Bradová, J., & Prášil, I. T. (2017). The effect of fusarium culmorum infection and deoxynivalenol (don) application on proteome response in barley cultivars chevron and pedant. Journal of Proteomics, 169, 112–124.
Kumar, Y., Zhang, L., Panigrahi, P., Dholakia, B. B., Dewangan, V., Chavan, S. G., et al. (2016). Fusarium oxysporum mediates systems metabolic reprogramming of chickpea roots as revealed by a combination of proteomics and metabolomics. Plant Biotechnology Journal, 14, 1589–1603.
Li, J., Pan, Y., & Liu, G. (2013). Disruption of the nitrogen regulatory gene acarea in acremonium chrysogenum leads to reduction of cephalosporin production and repression of nitrogen metabolism. Fungal Genetics and Biology, 61, 69–79.
López-Berges, M. S., Schäfer, K., Hera, C., & Di Pietro, A. (2014). Combinatorial function of velvet and area in transcriptional regulation of nitrate utilization and secondary metabolism. Fungal Genetics and Biology, 62, 78–84.
López-Díaz, C., Rahjoo, V., Sulyok, M., Ghionna, V., Martín-Vicente, A., Capilla, J., Di Pietro, A., & López-Berges, M. S. (2017). Fusaric acid contributes to virulence of fusarium oxysporum on plant and mammalian hosts. Molecular Plant Pathology. https://doi.org/10.1111/mpp.12536.
Luo, F., Zhong, Z., Liu, L., Igarashi, Y., Xie, D., & Li, N. (2017). Metabolomic differential analysis of interspecific interactions among white rot fungi trametes versicolor, dichomitus squalens and pleurotus ostreatus. Scientific Reports, 7, 5265.
Michielse, C. B., & Rep, M. (2009). Pathogen profile update: Fusarium oxysporum. Molecular Plant Pathology, 10, 311–324.
Nazari, F., Sulyok, M., Kobarfard, F., Yazdanpanah, H., & Krska, R. (2015). Evaluation of emerging fusarium mycotoxins beauvericin, enniatins, fusaproliferin and moniliformin in domestic rice in iran. Iranian Journal of Pharmaceutical Research: IJPR, 14, 505–512.
Oxoid. (2017a). Czapek dox agar (modified): Cm0097. In Dehydrated Culture Media. Walthman: Thermo Fisher Scientific Inc.
Oxoid. (2017b). Potato dextrose agar (ep/usp/jp/bp). In Dehydrated Culture Media. Walthman: Thermo Fisher Scientific Inc..
Oxoid. (2017c). Sabouraud dextrose agar code: Cm0041. In Dehydrated Culture Media. Walthman: Thermo Fisher Scientific Inc..
Panagiotou, G., Villas-Bôas, S. G., Christakopoulos, P., Nielsen, J., & Olsson, L. (2005). Intracellular metabolite profiling of Fusarium oxysporum converting glucose to ethanol. Journal of Biotechnology, 115, 425–434.
Pfannmüller, A., Leufken, J., Studt, L., Michielse, C. B., Sieber, C. M. K., Güldener, U., et al. (2017). Comparative transcriptome and proteome analysis reveals a global impact of the nitrogen regulators area and areb on secondary metabolism in fusarium fujikuroi. PLoS ONE, 12, e0176194.
Romero, F. M., Marina, M., Pieckenstain, F. L., Rossi, F. R., Gonzalez, M. E., Vignatti, P., & Gárriz, A. (2017). Gaining insight into plant responses to beneficial and pathogenic microorganisms using metabolomic and transcriptomic approaches. In V. C. Kalia & A. K. Saini (Eds.), Metabolic engineering for bioactive compounds: Strategies and processes (pp. 113–140). Singapore: Springer.
Sazanova, K. V., Vlasov, D. Y., Osmolovskay, N. G., Schiparev, S. M., & Rusakov, A. V. (2016). Significance and regulation of acids production by rock-inhabited fungi. In O. V. Frank-Kamenetskaya, E. G. Panova & D. Y. Vlasov (Eds.), Biogenic—abiogenic interactions in natural and anthropogenic systems (pp. 379–392). Cham: Springer.
Smith, C. A., O’Maille, G., Want, E. J., Qin, C., Trauger, S. A., Brandon, T. R., Custodio, D. E., Abagyan, R., & Siuzdak, G. (2005). Metlin: A metabolite mass spectral database. Therapeutic Drug Monitoring, 27, 747–751.
Steiber, A., Kerner, J., & Hoppel, C. L. (2004). Carnitine: A nutritional, biosynthetic, and functional perspective. Molecular Aspects of Medicine, 25, 455–473.
Sumner, L. W., Amberg, A., Barrett, D., Beale, M. H., Beger, R., Daykin, C. A., et al. (2007). Proposed minimum reporting standards for chemical analysis. Metabolomics, 3, 211–221.
Tan, K.-C., & Oliver, R. P. (2014). 12 metabolomics and proteomics to dissect fungal phytopathogenicity. In M. Nowrousian (Ed.), Fungal genomics (pp. 301–319). Berlin Heidelberg: Springer.
Vanella, A., Russo, A., Acquaviva, R., Campisi, A., Di Giacomo, C., Sorrenti, V., & Barcellona, M. L. (2000). L-propionyl-carnitine as superoxide scavenger, antioxidant, and DNA cleavage protector. Cell Biology and Toxicology, 16, 99–104.
Wishart, D. S., Jewison, T., Guo, A. C., Wilson, M., Knox, C., Liu, Y., et al. (2013). Hmdb 3.0—the human metabolome database in 2013. Nucleic Acids Research, 41, D801–D807.
Xia, J., & Wishart, D. S. (2016). Using metaboanalyst 3.0 for comprehensive metabolomics data analysis. Current Protocols in Bioinformatics, 55, 14.10.1–14.10.91.
Author information
Authors and Affiliations
Contributions
AVK and MSD performed the data analysis and co-wrote the paper; MCT performed the LC and GC analysis; TN, CO, and TK isolated the fungi and carried out the fungal experiments; SR and DJB devised and supervised the project. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Research involving human and animal participants
This article does not contain any studies with human and/or animal participants performed by any of the authors.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Karpe, A.V., Dunn, M.S., Taylor, M.C. et al. Nitrogen deprivation in Fusarium oxysporum promotes mycotoxin production via intermediates in the Krebs cycle and unreported methylmalonyl-CoA mutase activity. Metabolomics 14, 160 (2018). https://doi.org/10.1007/s11306-018-1459-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11306-018-1459-0