Abstract
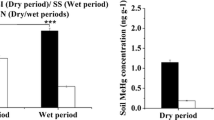
Sulfate-reducing microorganisms (SRM) have been thought to play a key role in mercury (Hg) methylation in anoxic environments. The current study examined the linkage between SRM abundance and diversity and contents of methylmercury (MeHg) in paddy soils collected from a historical Hg mining area in China. Soil profile samples were collected from four sites over a distance gradient downstream the Hg mining operation. Results showed that MeHg content in the soil of each site significantly decreased with the extending distance away from Hg mine. Soil MeHg content was correlated positively with abundance of SRM and the contents of organic matter (OM), NH4 +, SO4 2−, and Hg. The abundances of SRM based on dissimilatory (bi) sulfite reductase (dsrAB) gene at 0–40 cm depths were higher than those at 40–80 cm depth at all sites. The SRM community composition varied in the soils of different sampling sites following terminal restriction fragment length polymorphism (T-RFLP) and phylogenetic analyses, which appeared to be correlated with contents of MeHg, OM, NH4 +, and SO4 2− through canonical correspondence analysis. The dominant groups of SRM in the soils examined belonged to Deltaproteobacteria and some unknown SRM clusters that could have potential for Hg methylation. These results advance our understanding of the relationship between SRM and methylmercury concentration in paddy soil.







Similar content being viewed by others
References
Acha D, Iniguez V, Roulet M, Guimaraes JR, Luna R, Alanoca L, Sanchez S (2005) Sulfate-reducing bacteria in floating macrophyte rhizospheres from an Amazonian floodplain lake in Bolivia and their association with Hg methylation. Appl Environ Microbiol 71:7531–7535
Barkay T, Miller SM, Summers AO (2003) Bacteria mercury resistance from atoms to ecosystems. FEMS Microbiol Rev 27:355–384
Benoit JM, Gilmour CC, Mason RP (2001) Aspects of bioavailability of mercury for methylation in pure cultures of Desulfobulbus propionicus (1pr3). Appl Environ Microbiol 67:51–58
Branfireun BA, Roulet NT, Kelly CA, Rudd JWM (1999) In situ sulphate stimulation of mercury methylation in a boreal peatland: Toward a link between acid rain and methylmercury contamination in remote environments. Glob Biogeochem Cycles 13:743–750
Cao P, Zhang LM, Shen JP, Zheng YM, Di HJ, He JZ (2012) Distribution and diversity of archaeal communities in selected Chinese soils. FEMS Microbiol Ecol 80:146–158
Castro H, Reddy KR, Ogram A (2002) Composition and function of sulfate-reducing prokaryotes in eutrophic and pristine areas of the Florida Everglades. Appl Environ Microbiol 68:6129–6137
China National Environmental Monitoring Station (CNEMS) (1990) Background elemental concentrations of Chinese soils. Environmental Science Press, Beijing, China (in Chinese)
Choi SC, Chase T, Bartha R (1994) Metabolic pathways leading to mercuty in Desulfovibrio-desulfuricans LS. Appl Environ Microbiol 60:4072–4077
Coates JD, Anderson RT, Lovley DR (1996) Oxidation of polycyclic aromatic hydrocarbons under sulfate-reducing conditions. Appl Environ Microbiol 62:1099–1101
Compeau GC, Bartha R (1985) Sulfate-reducing bacteria: principal methylators of mercury in anoxic estuarine sediment. Appl Environ Microbiol 50:498–502
Dhillon A, Teske A, Dillon J, Stahl DA, Sogin ML (2003) Molecular characterization of sulfate-reducing bacteria in the Guaymas Basin. Appl Environ Microbiol 69:2765–2772
Duran R, Ranchou-Peyruse M, Menuet V, Monperrus M, Bareille G, Goni MS, Salvado JC, Amouroux D, Guyoneaud R, Donard OFX, Caumette P (2008) Mercury methylation by a microbial community from sediments of the Adour Estuary (Bay of Biscay, France). Environ Pollut 156:951–958
Ekstrom EB, Morel FMM, Benoit JM (2003) Mercury methylation independent of the acetyl-coenzyme a pathway in sulfate-reducing bacteria. Appl Environ Microbiol 69:5414–5422
Feng X, Li P, Qiu G, Wang S, Li G, Shang L, Meng B, Jiang H, Bai W, Li Z, Fu X (2008) Human exposure to methylmercury through rice intake in mercury mining areas, guizhou province, China. Environ Sci Technol 42:326–332
Fishbain S, Dillon JG, Gough HL, Stahl DA (2003) Linkage of high rates of sulfate reduction in Yellowstone hot springs to unique sequence types in the dissimilatory sulfate respiration pathway. Appl Environ Microbiol 69:3663–3667
Fitzgerald WF, Lamborg CH, Hammerschmidt CR (2007) Marine biogeochemical cycling of mercury. Chem Rev 107:641–662
Geets J, Borremans B, Diels L, Springael D, Vangronsveld J, Lelie DVD, Vanbroekhoven K (2006) DsrB gene-based DGGE for community and diversity surveys of sulfate-reducing bacteria. J Microbiol Methods 66:194–205
Gilmour CC, Henry EA, Mitchell R (1992) Sulfate stimulation of mercury methylation fresh-water sediments. Environ Sci Technol 26:2281–2287
Han S, Narasingarao P, Obraztsova A, Gieskes J, Hartmann AC, Tebo BM, Allen EE, Deheyn DD (2010) Mercury speciation in marine sediments under sulfate-limited conditions. Environ Sci Technol 44:3752–3757
He JZ, Liu XZ, Zheng Y, Shen JP, Zhang LM (2010) Dynamics of sulfate reduction and sulfate-reducing prokaryotes in anaerobic paddy soil amended with rice straw. Biol Fertil Soils 46:283–291
He JZ, Shen JP, Zhang LM, Zhu YG, Zheng YM, Xu MG, Di HJ (2007) Quantitative analyses of the abundance and composition of ammonia-oxidizing bacteria and ammonia-oxidizing archaea of a Chinese upland red soil under long-term fertilization practices. Environ Microbiol 9:3152–3152
Hsu-Kim H, Kucharzyk KH, Zhang T, Deshusses MA (2013) Mechanisms Regulating Mercury bioavailability for methylating microorganisms in the aquatic environment: a critical review. Environ Sci Technol 47:2441–2456
Heyes A, Mason RP, Kim EH, Sunderland E (2006) Mercury methylation in estuaries: Insights from using measuring rates using stable mercury isotopes. Mar Chem 102:134–147
Jiang L, Zheng Y, Peng X, Zhou H, Zhang C, Xiao X, Wang F (2009) Vertical distribution and diversity of sulfate-reducing prokaryotes in the Pearl River estuarine sediments, Southern China. FEMS Microbiol Ecol 70:249–262
Khwaja AR, Bloom PR, Brezonik PL (2010) Binding Strength of Methylmercury to Aquatic NOM. Environ Sci Technol 44:6151–6156
King JK, Kostka JE, Frischer ME, Saunders FM (2000) Sulfate-reducing bacteria methylate mercury at variable rates in pure culture and in marine sediments. Appl Environ Microbiol 66:2430–2437
Leloup J, Quillet L, Berthe T, Petit F (2006) Diversity of the dsrAB (dissimilatory sulfite reductase) gene sequences retrieved from two contrasting mudflats of the Seine estuary, France. FEMS Microbiol Ecol 55:230–238
Leloup J, Loy A, Knab NJ, Borowski C, Wagner M, Jorgensen BB (2007) Diversity and abundance of sulfate-reducing microorganisms in the sulfate and methane zones of a marine sediment, Black Sea. Environ Microbiol 9:131–142
Leloup J, Fossing H, Kohls K, Holmkvist L, Borowski C, Jorgensen BB (2009) Sulfate-reducing bacteria in marine sediment (Aarhus Bay, Denmark): abundance and diversity related to geochemical zonation. Environ Microbiol 11:1278–1291
Li JJ, Zheng YM, Yan JX, Li HJ, He JZ (2013) Succession of plant and soil microbial communities with restoration of abandoned land in the Loess Plateau, China. J Soils Sediments 13:760–769
Liang L, Horvat M, Feng XB, Shang LH, Li H, Pang P (2004) Re-evaluation of distillation and comparison with HNO3 leaching/solvent extraction for isolation of methylmercury compounds from sediment/soil samples. Appl Organomet Chem 18:264–270
Lin H, Shi J, Chen X, Yang J, Chen Y, Zhao Y, Hu T (2010) Effects of lead upon the actions of sulfate-reducing bacteria in the rice rhizosphere. Soil Biol Biochem 42:1038–1044
Liu XZ, Zhang LM, Prosser JI, He JZ (2009) Abundance and community structure of sulfate reducing prokaryotes in a paddy soil of southern China under different fertilization regimes. Soil Biol Biochem 41:687–694
Liu YR, Zheng YM, Shen JP, Zhang LM, He JZ (2010) Effects of mercury on the activity and community composition of soil ammonia oxidizers. Environ Sci Pollut Res 17:1237–1244
Liu YR, He JZ, Zhang LM, Zheng YM (2012) Effects of long-term fertilization on the diversity of bacterial mercuric reductase gene in a Chinese upland soil. J Basic Microbiol 52:35–42
Meng B, Feng XB, Qiu GL, Cai Y, Wang DY, Li P, Shang LH, Sommar J (2010) Distribution patterns of inorganic mercury and methylmercury in tissues of rice (Oryza sativa L.) plants and possible bioaccumulation pathways. J Agric Food Chem 58:4951–4958
Pak KR, Bartha R (1998) Mercury methylation by interspecies hydrogen and acetate transfer between sulfidogens and methanogens. Nucleic Acids Res 64:1987–1990
Parks JM, Johs A, Podar M, Bridou R, Hurt RA Jr, Smith SD, Tomanicek SJ, Qian Y, Brown SD, Brandt CC, Palumbo AV, Smith JC, Wall JD, Elias DA, Liang L (2013) The Genetic Basis for Bacterial Mercury Methylation. Science 339:1332–1335
Ranchou-Peyruse M, Monperrus M, Bridou R, Duran R, Amouroux D, Salvado JC, Guyoneaud R (2009) Overview of mercury methylation capacities among anaerobic bacteria including representatives of the sulphate-reducers: implications for environmental studies. Geomicrobiol J 26:1–8
Schaefer JK, Rocks SS, Zheng W, Liang L, Gu B, Morel FMM (2011) Active transport, substrate specificity, and methylation of Hg(II) in anaerobic bacteria. Proc Natl Acad Sci USA 108:8714–8719
Scheid D, Stubner S (2001) Structure and diversity of Gramnegative sulfate-reducing bacteria on rice roots. FEMS Microbiol Ecol 36:175–183
Schmalenberger A, Drake HL, Kusel K (2007) High unique diversity of sulfate-reducing prokaryotes characterized in a depth gradient in an acidic fen. Environ Microbiol 9:1317–1328
Sitte J, Akob DM, Kaufmann C, Finster K, BanerjeeD BEM, Kostka JE, Scheinost AC, Buechel G, Kuesel K (2010) Microbial Links between sulfate reduction and metal retention in uranium- and heavy metal-contaminated soil. Appl Environ Microbiol 76:3143–3152
Smith CJ, Nedwell DB, Dong LF, Osborn AM (2006) Evaluation of quantitative polymerase chain reaction based approaches for determining gene copy and gene transcript numbers in environmental samples. Environ Microbiol 8:804–815
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Todorova SG, Driscoll CT Jr, Matthews DA, Effler SW, Hines ME, Henry EA (2009) Evidence for Regulation of Monomethyl Mercury by Nitrate in a Seasonally Stratified, Eutrophic Lake. Environ Sci Technol 43:6572–6578
Walkley A, Black IA (1934) An examination of the Degtjareff method for determining soil organic matter, and a proposed modification of the chromic acid titration method. Soil Sci 37:29–38
Yu RQ, Adatto I, Montesdeoca MR, Driscoll CT, Hines ME, Barkay T (2010) Mercury methylation in Sphagnum moss mats and its association with sulfate-reducing bacteria in an acidic Adirondack forest lake wetland. FEMS Microbiol Ecol 74:655–668
Yu RQ, Flanders JR, Mack EE, Turner R, Mirza MB, Barkay T (2012) Contribution of coexisting sulfate and iron reducing bacteria to methylmercury production in freshwater river sediments. Environ Sci Technol 46:2684–2691
Zhang H, Feng XB, Larssen T, Shang L, Li P (2010) Bioaccumulation of methylmercury versus inorganic mercury in rice (Oryza sativa L.) grain. Environ Sci Technol 44:4499–450
Zhou ZF, Zheng YM, Shen JP, Zhang LM, Liu YR, He JZ (2012) Responses of activities, abundances and community structures of soil denitrifiers to short-term mercury stress. J Environ Sci 24:369–375
Acknowledgments
This work was supported by the National Natural Science Foundation of China (41201523, 41090281). We would like to thank Professor Peter Mather for improving English and Mr. Shi Xingwang for assistance in soil sampling.
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible editor: Robert Duran
Rights and permissions
About this article
Cite this article
Liu, YR., Zheng, YM., Zhang, LM. et al. Linkage between community diversity of sulfate-reducing microorganisms and methylmercury concentration in paddy soil. Environ Sci Pollut Res 21, 1339–1348 (2014). https://doi.org/10.1007/s11356-013-1973-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11356-013-1973-6