Abstract
Purpose
The purpose of this study was to determine the first-order rate constants and half-lives of aerobic and anaerobic biomineralization of atrazine in soil samples from an agricultural farm site that had been previously used for mixing pesticide formulations and washing application equipment. Atrazine catabolic genes and atrazine-degrading bacteria in the soil samples were analyzed by molecular methods.
Materials and methods
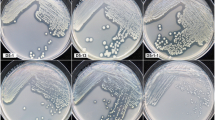
Biomineralization of atrazine was measured in soil samples with a [U-ring-14C]-atrazine biometer technique in soil samples. Enrichment cultures growing with atrazine were derived from soil samples and they were analyzed for bacterial diversity by constructing 16S rDNA clone libraries and sequencing. Bacterial isolates were also obtained and they were screened for atrazine catabolic genes.
Results and discussion
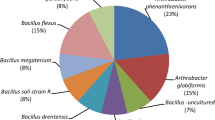
The soils contained active atrazine-metabolizing microbial communities and both aerobic and anaerobic biomineralization of [U-ring-14C]-atrazine to 14CO2 was demonstrated. In contrast to aerobic incubations, anaerobic biometers displayed considerable differences in the kinetics of atrazine mineralization between duplicates. Sequence analysis of 16S rDNA clone libraries constructed from the enrichment cultures revealed a preponderance of Variovorax spp. (51 %) and Schlesneria (16 %). Analysis of 16S rRNA gene sequences from pure cultures (n = 12) isolated from enrichment cultures yielded almost exclusively Arthrobacter spp. (83 %; 10/12 isolates). PCR screening of pure culture isolates for atrazine catabolic genes detected atzB, atzC, trzD, trzN, and possibly atzA. The presence of a complete metabolic pathway was not demonstrated by the amplification of catabolic genes among these isolates.
Conclusions
The soils contained active atrazine-metabolizing microbial communities. The anaerobic biometer data showed variable response of atrazine biomineralization to external electron acceptor conditions. Partial pathways are inevitable in soil microbial communities, with metabolites linking into other catabolic and assimilative pathways of carbon and nitrogen. There was no evidence for the complete set of functional genes of the known pathways of atrazine biomineralization among the isolates.





Similar content being viewed by others
References
Abigail MEA, Lakshmi V, Das N (2012) Biodegradation of atrazine by Cryptococcus laurentii isolated from contaminated agricultural soil. J Microbiol Biotechnol Res 2:450–457
Abigail MEA, Salam JA, Das N (2013) Atrazine degradation in liquid culture and soil by a novel yeast Pichia kudriavzevii strain Atz-EN-01 and its potential application for bioremediation. J Appl Pharmaceut Sci 3(6):35–40
Arthur EL, Perkovich BS, Anderson TA, Coats JR (2000) Degradation of an atrazine and metolachlor herbicide mixture in pesticide-contaminated soils from two agrochemical dealerships in Iowa. Water Air Soil Pollut 119:75–90
Aislabie J, Bej AK, Ryburn J, Lloyd N, Wilkins A (2005) Characterization of Arthrobacter nicotinovorans HIM, an atrazine-degrading bacterium, from agricultural soil New Zealand. FEMS Microbiol Ecol 52:279–286
Bastos AC, Magan N (2009) Trametes versicolor: potential for atrazine bioremediation in calcareous clay soil, under low water availability conditions. Int Biodeter Biodegr 63:389–394
Boundy-Mills KL, de Souza ML, Mandelbaum RT, Wackett LP, Sadowsky MJ (1997) The atzB gene of Pseudomonas sp. strain ADP encodes the second enzyme of a novel atrazine degradation pathway. Appl Environ Microbiol 63:916–923
Cheng G, Shapir N, Sadowsky MJ, Wackett LP (2005) Allophanate hydrolase, not urease, functions in bacterial cyanuric acid metabolism. Appl Environ Microbiol 71:4437–4445
Chirnside AEM, Ritter WF, Radosevich M (2007) Isolation of a selected microbial consortium from a pesticide-contaminated mix-load site soil capable of degrading the herbicides atrazine and alachlor. Soil Biol Biochem 39:3056–3065
Chirnside AEM, Ritter WF, Radosevich M (2009) Biodegradation of aged residues of atrazine and alachlor in a mixed-load site soil. Soil Biol Biochem 41:2484–2492
de Souza ML, Wackett LP, Boundy-Mills KL, Mandelbaum RT, Sadowsky MJ (1995) Cloning, characterization, and expression of a gene region from Pseudomonas sp. strain ADP involved in the dechlorination of atrazine. Appl Environ Microbiol 61:3373–3378
de Souza ML, Seffernick J, Martinez B, Sadowsky MJ, Wackett LP (1998) The atrazine catabolism genes atzABC are widespread and highly conserved. J Bacteriol 180:1951–1954
de Wilde T, Spanoghe P, Debaer C, Ryckeboer J, Springael D, Jaeken P (2007) Overview of on-farm bioremediation systems to reduce the occurrence of point source contamination. Pest Manag Sci 63:111–128
del Pilar Castillo M, Torstensson L, Stenström J (2008) Biobeds for environmental protection from pesticide use—a review. J Agric Food Chem 56:6206–6219
Devers M, Soulas G, Martin-Laurent F (2004) Real-time reverse transcription PCR analysis of expression of atrazine catabolism genes in two bacterial strains isolated from soil. J Microbiol Meth 56:3–15
Devers M, El Azhari N, Kolic N-U, Martin-Laurent F (2007) Detection and organization of atrazine-degrading genetic potential of seventeen bacterial isolates belonging to divergent taxa indicate a recent common origin of their catabolic functions. FEMS Microbiol Lett 273:78–86
Donnelly PK, Entry JA, Crawford DL (1993) Degradation of atrazine and 2,4-dichlorophenoxyacetic acid by mycorrhizal fungi at three nitrogen concentrations in vitro. Appl Environ Microbiol 59:2642–2647
Douglass JF, Radosevich M, Tuovinen OH (2014) Mineralization of atrazine in the river water intake and sediments of a constructed flow-through wetland. Ecol Eng 72:35–39
Douglass JF, Radosevich M, Tuovinen OH (2015) Molecular analysis of atrazine-degrading bacteria and catabolic genes in the water column and sediment of a created wetland in an agricultural/urban watershed. Ecol Eng 83:405–412
Eaton RW, Karns JS (1991) Cloning and analysis of s-triazine catabolic genes from Pseudomonas sp. strain NRRLB-12227. J Bacteriol 173:1215–1222
El Sebaï T, Devers-Lamrani M, Changey F, Rouard N, Martin-Laurent F (2011) Evidence of atrazine mineralization in a soil from the Nile Delta: isolation of Arthrobacter sp. TES6, an atrazine-degrading strain. Int Biodeter Biodegrad 65:1249–1255
Ettema CH, Wardle DA (2002) Spatial soil ecology. Trends Ecol Evol 17:177–183
Fan X, Song F (2014) Bioremediation of atrazine: recent advances and promises. J Soils Sedim 14:1727–1737
Fenner K, Canonica S, Wackett LP, Elsner M (2013) Evaluating pesticide degradation in the environment: blind spots and emerging opportunities. Science 341:752–758
Ghosh D, Roy K, Srinivasan V, Mueller T, Tuovinen OH, Sublette K, Peacock A, Radosevich M (2009) In-situ enrichment and analysis of atrazine-degrading microbial communities using atrazine-containing porous beads. Soil Biol Biochem 41:1331–1334
Grigg BC, Assaf NA, Turco RF (1997) Removal of atrazine contamination in soil and liquid systems using bioaugmentation. Pestic Sci 50:211–220
Guerin WF, Boyd SA (1992) Differential bioavailability of soil-sorbed naphthalene to two bacterial species. Appl Environ Microbiol 58:1142–1152
Karns JS, Eaton RW (1997) Genes encoding s-triazine degradation are plasmid-borne in Klebsiella pneumoniae strain 99. J Agric Food Chem 45:1017–1022
Krutz LJ, Shaner DL, Weaver MA, Webb RMT, Zablotowicz RM, Reddy KN, Huang Y, Thomson SJ (2010) Agronomic and environmental implications of enhanced s-triazine degradation. Pest Manag Sci 66:461–481
Krutz LJ, Zablotowicz RM, Reddy KN (2012) Selection pressure, cropping system, and rhizosphere proximity affect atrazine degrader populations and activity in s-triazine-adapted soil. Weed Sci 60:516–524
Kulichevskaya IS, Ivanova AO, Belova SE, Baulina OI, Bodelier PLE, Rijpstra WIC, Damsté JSS, Zavarzin GA, Dedysh SN (2007) Schlesneria paludicola gen. nov., sp. nov., the first acidophilic member of the order Planctomycetales, from Sphagnum-dominated boreal wetlands. Int J System Evol Microbiol 57:2680–2687
Lane DJ (1991) 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M (eds) Nucleic Acid Techniques in Bacterial Systematics. Wiley, Chichester, pp 115–175
Lazorko-Connon S, Achari G (2009) Atrazine: its occurrence and treatment in water. Environ Rev 17:199–214
Lian B, Jiang J, Zhang J, Zhao Y, Li S (2012) Horizontal transfer of dehalogenase genes involved in the catalysis of chlorinated compounds: evidence and ecological role. Crit Rev Microbiol 38:95–110
Liu X, Hui C, Bi L, Romantschuk M, Kontro M, Strömmer R, Hui N (2016) Bacterial community structure in atrazine treated reforested farmland in Wuying China. Appl Soil Ecol 98:39–46
Martinez B, Tomkins J, Wackett LP, Wing R, Sadowsky MJ (2001) Complete nucleotide sequence and organization of the atrazine catabolic plasmid pADP-1 from Pseudomonas sp. strain ADP. J Bacteriol 183:5684–5697
Masaphy S, Levanon D, Henis Y (1996) Degradation of atrazine by the lignocellulolytic fungus Pleurotus pulmonarius during solid-state fermentation. Bioresour Technol 56:207–214
Mougin C, Laugero C, Asther M, Dubroca J, Frasse P, Asther M (1994) Biotransformation of the herbicide atrazine by the white rot fungus Phanerochaete chrysosporium. Appl Environ Microbiol 60:705–708
Mudhoo A, Garg VK (2011) Sorption, transport and transformation of atrazine in soils, minerals and composts: a review. Pedosphere 21:11–25
Mueller TC, Steckel LE, Radosevich M (2010) Effect of soil pH and previous atrazine use history on atrazine degradation in a Tennessee field soil. Weed Sci 58:478–483
Mulbry WW, Zhu H, Nour SM, Topp E (2002) The triazine hydrolase gene trzN from Nocardioides sp. strain C190: cloning and construction of gene-specific primers. FEMS Microbiol Lett 206:75–79
Muyzer G, de Waal EC, Uitterlinden AG (1993) Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol 59:695–700
O’Donnell AG, Young IM, Rushton SP, Shirley MD, Crawford JW (2007) Visualization, modelling and prediction in soil microbiology. Nature Rev Microbiol 5:689–699
Omotayo AE, Ilori MO, Amund OO, Ghosh D, Roy K, Radosevich M (2011) Establishment and characterization of atrazine degrading cultures from Nigerian agricultural soil using traditional and Bio-Sep bead enrichment techniques. Appl Soil Ecol 48:63–70
Pussemier L, Goux S, Vanderheyden V, Debongnie P, Tresinie I, Foucart G (1997) Rapid dissipation of atrazine in soils taken from various maize fields. Weed Res 37:171–179
Radosevich M, Traina SJ, Hao Y-L, Tuovinen OH (1995) Degradation and mineralization of atrazine by a soil bacterial isolate. Appl Environ Microbiol 61:297–302
Radosevich M, Traina SJ, Tuovinen OH (1996) Biodegradation of atrazine in surface soils and subsurface sediments collected from an agricultural research farm. Biodegradation 7:137–149
Romero MC, Urrutia MI, Reinoso EH, Vedoval RD, Reynaldi FJ (2014) Atrazine degradation by wild filamentous fungi. Global Res J Microbiol 4:10–16
Rousseaux S, Hartmann A, Soulas G (2001) Isolation and characterisation of new Gram-negative and Gram-positive atrazine degrading bacteria from different French soils. FEMS Microbiol Ecol 36:211–222
Sadowsky MJ, Tong Z, de Souza M, Wackett LP (1998) AtzC is a new member of the amidohydrolase protein superfamily and is homologous to other atrazine-metabolizing enzymes. J Bacteriol 180:152–158
Sajjaphan K, Shapir N, Wackett LP, Palmer M, Blackmon B, Tomkins J, Sadowsky MJ (2004) Arthrobacter aurescens TC1 atrazine catabolism genes trzN, atzB, and atzC are linked on a 160-kilobase region and are functional in Escherichia coli. Appl Environ Microbiol 70:4402–4407
Sajjaphan K, Heepngoen P, Sadowsky MJ, Boonkerd N (2010) Arthrobacter sp. strain KU001 isolated from a Thai soil degrades atrazine in the presence of inorganic nitrogen sources. J Microbiol Biotechnol 20:602–608
Singh B, Singh K (2016) Microbial degradation of herbicides. Crit Rev Microbiol 42:245–261
Smith D, Alvey S, Crowley DE (2005) Cooperative catabolic pathways within an atrazine-degrading enrichment culture isolated from soil. FEMS Microbiol Ecol 53:265–273
Spanoghe P, Maes A, Steurbaut W (2009) Limitation of point source pesticide pollution: results of bioremediation system. Comm Appl Biol Sci Ghent Univ 74(2):1–14
Stamper DM, Radosevich M, Hallberg KB, Traina SJ, Tuovinen OH (2002) Ralstonia basilensis M91-3, a denitrifying soil bacterium capable of using s-triazines as nitrogen sources. Can J Microbiol 48:1089–1098
Tuovinen OH, Desmukh V, Özkaya B, Radosevich M (2015) Kinetics of aerobic and anaerobic biomineralization of atrazine in surface and subsurface agricultural soils in Ohio. J Environ Sci Health B50:718–726
Udiković N, Hršak D, Mendaš G, Filipčić D (2003) Enrichment and characterization of atrazine degrading microbial communities. Food Technol Biotechnol 41:211–217
Udiković Kolić N, Hršak D, Kolar AB, Petrić I, Stipičevic S, Soulas G, Martin-Laurent F (2007) Combined metabolic activity within an atrazine-mineralizing community enriched from agrochemical factory soil. Int Biodeter Biodegrad 60:299–307
Udiković-Kolić N, Hršak D, Devers M, Klepac-Ceraj V, Petrič I, Martin-Laurent F (2010) Taxonomic and functional diversity of atrazine-degrading bacterial communities enriched from agrochemical factory soil. J Appl Microbiol 109:355–367
Udiković-Kolić N, Scott C, Martin-Laurent F (2012) Evolution of atrazine-degrading capabilities in the environment. Appl Microbiol Biotechnol 96:1175–1189
Vargha M, Takátsc Z, Márialigeti K (2005) Degradation of atrazine in a laboratory scale model system with Danube river sediment. Water Res 39:1560–1568
Vaishampayan PA, Kanekar PP, Dhakephalkar PK (2007) Isolation and characterization of Arthrobacter sp. strain MCM B-436, an atrazine-degrading bacterium, from rhizospheric soil. Int Biodeter Biodegr 60:273–278
Vibber LL, Pressler MJ, Colores GM (2007) Isolation and characterization of novel atrazine-degrading microorganisms from an agricultural soil. Appl Microbiol Biotechnol 75:921–928
Zhang Y, Jiang Z, Cao B, Hu M, Wang Z, Dong X (2011) Metabolic ability and gene characteristics of Arthrobacter sp. strain DNS10, the sole atrazine-degrading strain in a consortium isolated from black soil. Int Biodeter Biodegrad 65:1140–1144
Zhang Y, Cao B, Jiang Z, Dong X, Hu M, Wang Z (2012) Metabolic ability and individual characteristics of an atrazine-degrading consortium DNC5. J Hazard Mater 237–238:376–381
Acknowledgments
The work was funded through the USDA National Research Initiative, grant no. 2004-35107-14884. JFD gratefully acknowledges a Fellowship from the Robert H. Edgerley Environmental Toxicology Fund, Division of Natural and Mathematical Sciences of the College of Arts and Sciences, Ohio State University. We thank two anonymous reviewers for insightful suggestions that were helpful in improving the manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible editor: Jizheng He
Rights and permissions
About this article
Cite this article
Douglass, J.F., Radosevich, M. & Tuovinen, O.H. Biomineralization of atrazine and analysis of 16S rRNA and catabolic genes of atrazine degraders in a former pesticide mixing site and a machinery washing area. J Soils Sediments 16, 2263–2274 (2016). https://doi.org/10.1007/s11368-016-1416-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11368-016-1416-3