Abstract
Silicon (Si) enhances plant tolerance to various biotic and abiotic stressors such as salinity, drought, and heat. In addition, Si can be biomineralized within plants to form organic carbon-containing phytoliths that can have ecosystem-level consequences by contributing to long-term carbon sequestration. Si is taken up and transported in plants via different transporter proteins such as influx transporters (e.g., Lsi1, Lsi6) and efflux transporters (e.g., Lsi2). Additionally, the imported Si can be deposited in plant leaves via silicification process using the Siliplant 1 (e.g., Slp1) protein. Functional homologs of these proteins have been reported in different food crops. Here, we performed a genome-wide analysis to identify different Si transporters and Slp1 homologs in the bioenergy crop poplar (Populus trichocarpa Torr. and A. Gray ex W. Hook). We identified one channel-type Si influx transporter (PtLsi1; Potri.017G083300), one Si efflux transporter (PtLsi2; Potri.012G144000) and two proteins like Slp1 (PtSlp1a; Potri.004G168600 and PtSlp1b; Potri.009G129900) in the P. trichocarpa genome. We found a unique sequence (KPKPPVFKPPPVPI) in PtSlp1a which is repeated six times. Repeated presence of this sequence in PtSlp1a indicates that this protein might be important for silicification processes in P. trichocarpa. The mutation profiles of different Si transporters in a P. trichocarpa genome-wide association study population identified significant and impactful mutations in Potri.004G168600 and Potri.009G129900. Using a publically accessible database (http://bar.utoronto.ca/eplant_poplar/), digital expression analysis of the putative Si transporters in P. trichocarpa found low to moderate expression in the anticipated tissues, such as roots and leaves. Subcellular localization analysis found that PtLsi1/PtLsi2 are localized in the plasma membrane, whereas PtSlp1a/PtSlp1b are found in the extracellular spaces. Protein–Protein interaction analysis of PtLsi1/PtLsi2 identified Delta-1-pyrroline-5-carboxylate synthase (P5CS) as one of the main interacting partners of PtLsi2, which plays a key role in proline biosynthesis. Proline is a well-known participant in biotic and abiotic stress tolerance in plants. These findings will reinforce future efforts to modify Si accumulation for enhancing plant stress tolerance and carbon sequestration in poplar.
Similar content being viewed by others
Introduction
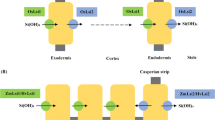
Silicon (Si) is the second-most abundant (28.8%) element in Earth’s continental crust after oxygen (Kaur and Greger 2019). The uptake and accumulation of Si in plants is associated with a number of benefits (Fig. 1). For example, Si enhances plant tolerance to various abiotic (e.g., drought, salinity, metal toxicity) and biotic (e.g., pathogen, herbivory) stressors (Souri et al. 2020; Gaur et al. 2020; Ahanger et al. 2020). The accumulation of Si in plants as carbon-containing phytoliths (Si-based structures in plant cells) also contributes to long-term carbon sequestration (Hans Wedepohl 1995). However, Si is considered a non-essential element for most plants. Plants acquire Si as silicic acid [Si(OH)4] from the soil through the root system. Silicic acid is then translocated to the aboveground tissues and organs and deposited as phytoliths through the process of biosilicification (Kaur and Greger 2019).
The uptake, transport and deposition of Si from soil to a plant is mediated by a cooperative action of various transporter genes, such as channel-type influx (e.g., Lsi1/Lsi6) and efflux (e.g., Lsi2) transporters (Kaur and Greger 2019; Nawaz et al. 2020). The first Si transporter gene identified in plants was rice (Oryza sativa L.) low silicon 1 (OsLsi1) (Ma et al. 2006). Functional homologs of OsLsi1 have since been identified in many plant species, including in barley (Hordeum vulgare L.) (Chiba et al. 2009), pumpkin (Cucurbita moschata Duch.) (Mitani et al. 2011), wheat (Triticum aestivum L.) (Montpetit et al. 2012), soybean (Glycine max L.) (Deshmukh et al. 2013), maize (Zea mays L.) (Mitani et al. 2009b) and cucumber (Cucumis sativus L.) (Sun et al. 2017). Lsi1 belongs to the Nodulin 26-like intrinsic protein (NIP) III subgroup of the aquaporin gene family (Ma and Yamaji 2015). This gene is involved in Si uptake from the soil into root cells in both eudicots and monocots. Functional characterization of the Lsi1 protein in different plant species shows that Si specificity in plants is linked to the two highly conserved Asn-Pro-Ala (NPA) motifs and an aromatic/arginine (ar/R) region formed by four amino acid (AA) residues that act as a selectivity filter (SF). The SFs consist of Gly-Ser-Gly-Arg (GSGR) or Ser-Thr-Ala-Arg (STAR) residues (Mitani-Ueno et al. 2011b; Grégoire et al. 2012). In addition, a precise spacing of 108 AAs between the two NPA motifs is essential for Si permeability in plants (Deshmukh et al. 2015). Lsi2, an efflux transporter gene which was first identified in rice, transports Si from root cells to the root xylem tissue. It belongs to an uncharacterized putative anion transporter protein family. Homologs of Lsi2 have been reported in several plants, such as barley, maize (Mitani et al. 2009a) and pumpkin (Mitani-Ueno et al. 2011a).
Si in xylem sap is deposited as monosilicic acid which is then unloaded by Lsi6, a homolog of Lsi1 (Yamaji et al. 2008). Lsi6 subsequently distributes Si to the aboveground tissues where it is deposited as amorphous Si(SiO2) in phytoliths (Kumar et al. 2017). In some plants, there are specialized cells called silica cells (a leaf epidermal cell) where Si is deposited. The molecular mechanism of silica deposition has recently been uncovered in Sorghum with the identification of the protein Siliplant 1 (Slp1) (Kumar et al. 2017). Slp1 contains a short peptide (HKKPVPPKPKPEPK) that controls the silicification process in Sorghum (Kumar et al. 2020). A generalized scheme of Si uptake, transport and deposition in plants is depicted in Fig. 1.
The expression and subcellular localization of Lsi1 and Lsi2 in roots varies in different plant species, despite these transporters having similar functions. In rice roots, OsLsi1 is found on the distal side of root exodermal and endodermal cells (Ma et al. 2006, 2007). On the other hand, the maize homolog of OsLsi1, ZmLsi1, is found in seminal and crown roots on the distal side of root epidermal and hypodermal cells, as well as in the cortical cells of lateral roots (Mitani et al. 2009b). In barley, HvLsi1 is localized on the distal side of epidermal and cortical cells in seminal roots and on the distal side of hypodermal cells in lateral roots (Chiba et al. 2009). Like Lsi1, subcellular localization of Lsi2 varies by plant species. For example, in rice, OsLsi2 is localized to the proximal side of root exodermal and endodermal cells (Ma et al. 2006, 2007), while maize and barley, Lsi2 transporters are only localized to root endodermal cells and show no polar distribution (Mitani et al. 2009a). The difference in cellular localizations of Si transporters in different plant species may be associated with their Si uptake ability. Using a mathematical simulation model, Sakurai et al. (2015, 2017) concluded that the most efficient Si uptake strategy is with OsLsi1 and OsLsi2 localized to the exodermis and endodermis with a polar distribution. This model simulation is, therefore, predicting that rice a high Si accumulator. To test such predictions, Si transporters need to be experimentally identified and characterized across multiple plant species to better understand how localization affects Si uptake and accumulation.
As described above, previous studies on Si accumulation have focused on food crops (e.g., barley, rice, wheat, maize, soybean) (Souri et al. 2020; Ahanger et al. 2020). However, our knowledge on the genetic variability and molecular mechanisms of Si accumulation in bioenergy crops such as switchgrass (Panicum virgatum L.) and poplar (Populus trichocarpa Torr. & A. Gray ex W. Hook) is limited. While Si uptake by plants can have multiple benefits (Fig. 1), there may be negative impacts on the utilization of these crops for bioenergy production as Si in plant tissues with high Si content may inhibit the bioprocessing of bioenergy feedstocks. Thus, manipulating the phytolith-forming pathway, including uptake, transport, and translocation of Si in plants, will directly influence plant productivity and potentially extend to influence bioprocessing and ecosystem-level processes such as carbon sequestration (Fig. 1). In this study, we conducted an in-silico genome analysis to identify the putative transporter genes involved in Si uptake, transport, and deposition in poplar, a predominant bioenergy crop.
Materials and methods
Clustering of protein sequences into tribes
The protein-coding sequences (CDSs) of the primary transcripts annotated in the genomes of six plant species including Arabidopsis thaliana (L.) Heynh. (Araport11), Amborella trichopoda Baill. (v1.0), Oryza sativa L. (v7.0), Populus trichocarpa Torr. & A.Gray ex Hook. (v4.1), Sorghum bicolor (L.) Moench (v3.1.1), and Solanum lycopersicum L. (ITAG3.2) were obtained from Phytozome v13 (https://phytozome-next.jgi.doe.gov/) (Goodstein et al. 2012). The protein sequences translated from the CDSs of individual plant species were combined into a single protein sequence file in fasta format. Then 'all-against-all' sequence alignments in this combined protein sequence dataset were performed using DIAMOND BLASTP (Buchfink et al. 2015) with parameters of “-e 1e-5 -f 6 -k 1000000”. The BLASTP output was used for clustering the protein sequences into tribes using TRIBE-MCL (Enright et al. 2002), with various inflation values (i.e., 1.1, 1.2, 1.3, …, 2.0).
Sequence analysis
To identify Pfam domains (Bateman et al. 2002; Mistry et al. 2021), the protein sequences described in section “clustering of protein sequences into tribes”, were analyzed using InterProScan version 5.48–83.0 (Jones et al. 2014) with parameters of “-f tsv -appl Pfam”. For channel-type silicon-influx transporters (SITs), the SF and NPA motif were manually searched against OsLsi1. The distance between the two NPA motifs was manually calculated. Transmembrane domains (TMDs) were predicted by the TMHMM server (https://services.healthtech.dtu.dk/service.php?TMHMM-2.0) (Krogh et al. 2001) and the topology was visualized with the PROTTER (Omasits et al. 2014) tool. Theoretical isoelectric point (pI) and amino acid frequencies were computed using protein identification and analysis tools on the ExPASy Server (https://web.expasy.org/compute_pi/) (Gasteiger et al. 2005). Repeats in protein sequences were predicted using the RADAR web server (https://ebi.ac.uk/Tools/pfa/radar/) (Madeira et al. 2019). Conserved motifs were found in the peptide sequences using the MEME tool (https://meme.nbcr.net/meme/tools/meme) (Bailey et al. 2006). SignalP was used to predict signal peptide and cleavage sites (Armenteros et al. 2019).
Phylogenetic analysis
Phylogenetic analysis of protein sequences was performed as described previously (Yang et al. 2017). Specifically, protein sequences of the six plant species in each gene family containing known Si transporters were aligned using MAFFT (v7.453) (Katoh and Standley 2013) or CLC sequence viewer (http://www.qiagen.com), with the setting of “auto”. The protein sequence alignment was edited using Jalview software (Waterhouse et al. 2009). The protein sequence alignment for each gene family was used to infer maximum-likelihood phylogenetic tree using IQ-TREE 2 (version 2.1.2) (Minh et al. 2020), with the setting of “-B 1000”. Finally, the phylogenetic trees were visualized using MEGA X (version 10.1.6) (Kumar et al. 2018).
Gene ontology analysis and pathway annotation
Gene ontology (GO) analysis of protein sequences was performed using DeepGOPlus (version 1.0.1) (Kulmanov and Hoehndorf 2020), with a prediction threshold of 0.3. Pathway annotation of protein sequences in each gene family containing known Si transporters was performed on the KEGG Blast KOALA (Kanehisa et al. 2016), with the KEGG GENES database as “family_eukaryotes”.
Prediction of protein subcellular localization
The protein sequences in each gene family containing known Si transporters were analyzed for predicting subcellular localization using three different tools, including Plant-mSubP (http://bioinfo.usu.edu/Plant-mSubP/) (Sahu et al. 2020) with the prediction module set as “PseAACNCCDipep”, BUSCA (http://busca.biocomp.unibo.it/) with the taxonomic origin set as “Eukarya—Plants—16 compartments”, and DeepLoc-1.0 (https://services.healthtech.dtu.dk/service.php?DeepLoc-1.0) (Almagro Armenteros et al. 2017) with parameters set as Profiles (for 50 sequences or less) or BLOSUM62 (for more than 50 sequences).
Gene expression analysis
The gene expression data for the Si transporter genes in P. trichocarpa were obtained from ePlant Poplar (http://bar.utoronto.ca/eplant_poplar/) (Waese et al. 2017). Briefly, Affymetrix expression data were normalized by the GCOS method, with a TGT value of 500. Duplicate or triplicate samples were analyzed from greenhouse-grown or field-grown material (the latter in the case of the catkins). The seedings were grown on moist filter paper. All materials were grown under a diurnal cycle of 12 h light/12 h dark and sampled at midday, except for the xylem samples, which were sampled at midnight.
Protein 3D structure analysis
The three-dimensional (3D) structures of the Si transporter and silicification proteins were determined with AlphaFold 2 on Google Colab’s Notebook (Mirdita et al. 2022). This Colab notebook allows for the prediction of protein structure using a slightly simplified version of AlphaFold v.2.1.0. Colab notebooks are coupled to a special sequence search program, MMSeqs2, thus making this technology more powerful. The predicted structures were downloaded from Google Colab’s server in protein databank (PDB) format and visualized with the FirstGlance in Jmol server (http://firstglance.jmol.org).
Analysis of gene duplications
MCScanX (Wang et al. 2012) was used to detect gene synteny among different plant species as well as segmental duplication within species. The tandem gene duplication information was obtained from PLAZA 4.5 (Van Bel et al 2018).
Analysis of gene networks
The protein sequences of P. trichocarpa were used to search the UniProt database through BLAST (https://www.uniprot.org/blast/) to obtain the corresponding proteins with UniProt accession numbers. Then, the UniProt accession numbers were used as queries to search the STRING (Szklarczyk et al. 2021) database for genes connected to each query gene.
Genome-wide association studies (GWAS)
Mutation profiles for each gene were obtained from whole-genome resequencing of 917 unrelated P. trichocarpa genotypes that were used to assemble the GWAS as described by Muchero et al. (2018). Expression analysis for the GWAS panel was performed using RNA-seq as previously described (Zhang et al. 2018). In the work by Muchero et al. (2018) and Zhang et al. (2018), 533 xylem and 470 leaf samples were collected from 4-year-old trees from the Clatskanie field site, Oregon, USA, while 529 root samples were collected greenhouse-grown plants at Oak Ridge National Laboratory, Tennessee, USA.
Results and discussion
Identification of Si transporters in P. trichocarpa
Channel-type Si influx transporters (SITs)
Until now, two channel-type SIT genes (Lsi1/Lsi6) have been reported in different plant species such as rice, wheat, maize, barley and soybean (Souri et al. 2020). Here, we performed an in-silico analysis to identify the putative channel-type SITs in poplar. We first compared the protein coding genes of A. thaliana, A. trichopoda, O. sativa, P. trichocarpa, S. bicolor and S. lycopersicum at the whole-genome level to identify the putative channel-type SITs in poplar. Phylogenetic analysis of the whole-gene family containing the channel-type SITs (i.e., Lsi1/Lsi6) grouped them into four distinct clades (Fig. 2A). Further analysis of the subfamily containing the known SIT genes of rice (OsLsi1/OsLsi6) revealed that this clade contained 11 putative SITs in poplar (Fig. 2B). Out of the 11 putative SITs found in the poplar genome, only Potri.017G083300.1 (here referred to as PtLsi1) was grouped into the clade containing the rice SIT, indicating that this gene might be the candidate channel-type SIT in poplar. Further sequence analysis showed that PtLsi1 is ~ 55% similar to OsLsi1 and ~ 53% similar to OsLsi6 at the AA level.
Phylogenetic trees of the gene families containing different Si transporters. A The phylogenetic tree of the Lsi1/Lsi6 gene family, which was rooted on midpoint. A subfamily containing the known Si transporters OsLsi1 (LOC_Os02g51110) and OsLsi6 (LOC_Os06g12310) is highlighted in blue. B Phylogenetic tree of the subfamily containing the known Si transporters OsLsi1 (LOC_Os02g51110) and OsLsi6 (LOC_Os06g12310). The clade containing the two known Si transporters is highlighted in purple. C Phylogenetic tree of the gene family containing OsLsi2. The clade containing the known Si transporter OsLsi2 (LOC_Os03g01700) is highlighted in purple. D Phylogenetic tree of the gene family containing the known silicification protein SbSlp1 (Sobic.001G266300.1)
To examine the presence of NPA motifs and the SF in the putative PtLsi1, we aligned the AA sequences of rice channel-type SIT, i.e., OsLsi1/OsLsi6 with the putative PtLsi1 identified in this study. The alignment (Fig. 3A) shows that PtLsi1 contains both NPA motifs and SF, and their locations match the positions of NPA/SF reported in OsLsi1/OsLsi6. Similar to OsLsi1/OsLsi6, the distance between the two NPA motifs in PtLsi1 is 108 AA. The channel-type SITs in plants have six transmembrane domains (TMDs) (Wallace and Roberts 2004; Ma et al. 2006; Nawaz et al. 2020). We also found six TMDs in PtLsi1 (Fig. 3B), indicating that PtLsi1 is a membrane-bound protein and has an active role in transport across cells. Protein topology analysis showed that the N-terminus and C-terminus of PtLsi1 are located inside the intracellular space whereas in OsLsi1, these regions are in the extracellular space (Fig. 3B). Subcellular localization of PtLsi1 was determined using three different tools, including Wolfpsort (Horton et al. 2007), Cello (Yu et al 2006), and DeepLoc (Almagro Armenteros et al. 2017), and all tools predicted that PtLsi1 is a membrane-bound protein. A previous report in pumpkin (Mitani et al. 2011) showed that Si transport activity is hampered by a proline-to-leucin mutation in CmLsi1 at position 242 resulting in a mislocalization to the endoplasmic reticulum instead of the plasma membrane, which is essential for its activity. This mutation, however, was not found in PtLsi1.
Sequence analysis of a channel-type silicon influx transporter (SIT) in poplar. A Protein sequence alignment of the rice channel-type SIT (OsLsi1/OsLsi6) with the putative poplar homolog (PtLsi1) identified in this study. Signature motifs that characterize the channel-type SIT in plants are indicated by boxes, with the blue boxes showing the GSGR filter and the pink boxes showing the NPA motif. B Shows the presence of transmembrane domains in OsLsi1 and PtLsi1
A previous study (Vatansever et al. 2017) found that sequence similarity and Si transport ability can be used to infer accumulation capacities of different plant species. Applying a similar approach to our findings, it is likely that PtLsi1 is a high Si accumulator given sequence similarity to the high accumulating rice homolog (Vatansever et al. 2017). Indeed, a previous study (Deshmukh et al. 2015) identified “Potri.017G083300.1” as the candidate PtLsi1 in P. trichocarpa and showed that it can accumulate high levels of Si in Xenopus (clawed frog) eggs, similar to that observed with rice OsLsi1. However, when assessed in poplar plants, Deshmukh et al. (2015) found moderate levels of Si accumulation. This might be explained by the low expression of PtLsi1 in poplar roots (see section “Gene expression pattern of Si transporters in poplar” for details). One interesting observation is that the number of influx transporters is highly variable in different plant species. For example, rice contains two influx transporters (Lsi1, Lsi6) whereas maize has as many as four homologs, and both rice and maize accumulate high levels of Si. This is also the case for sunflower (Helianthus annuus L.) which accumulates Si and contains more than one channel-type influx transporter. Whether the number of Si transporters affects a plant species’ ability to take up Si has yet to be verified experimentally. Although rice has two Si influx transporter genes, only one homolog of Lsi1 was identified in P. trichocarpa. We did not find any homologs of Lsi6 in P. trichocarpa, which is involved in unloading Si from xylem and transporting it to multiple aboveground tissues. Interestingly, it has been shown that the absence of Lsi6 has no negative effect on Si uptake but can affect the pattern of Si deposition in leaves (Ma et al. 2011; Kaur and Greger 2019).
Si efflux transporter
Lsi2 is the only efflux transporter identified in plants to date. It belongs to an uncharacterized anion transporter family. Our genome-wide analysis identified 22 genes in this family, including five homologs in poplar (Potri.012G144000.5, Potri.015G146900.5, Potri.017G141600.3, Potri.017G 141800.2, Potri.017G141950.1). This phylogenetic analysis grouped the 22 genes into six distinct clades (Fig. 2C). The clade containing the rice Lsi2 (OsLsi2; LOC_Os02g51110) harbors only one poplar gene Potri.012G144000 (here referred to as PtLsi2). We next aligned the protein sequences of five poplar homologs along with OsLsi2 to visualize their conserved regions (Fig. 4A). The alignments showed that these genes are highly conserved over the full-length of the protein sequences except at positions 230–375. The similarity between the OsLsi2 and the putative PtLsi2 is ~ 50% at the AA level. We found five conserved motifs in the putative Lsi2s (except Potri.017G141950.1) in poplar and in OsLsi2 (Fig. 4B).
Sequence alignment and motif analysis of putative Lsi2 homologs in poplar. A Sequence alignment of the putative Lsi2 homolog in poplar identified in our genome-wide study. Transmembrane domains (TMDs) are highlighted with shaded colors. Each different color indicates a different TMD. B Organization of the five highly conserved motifs predicted in the putative Lsi2 homolog in poplar by the MEME tool
Previous studies found that the Lsi2 protein in plants is localized in the plasma membrane, featuring 9–11 TMDs, with extracellular N- and extracellular/cytoplasmic C-terminus regions (Vivancos et al. 2016; Vatansever et al. 2017; Nawaz et al. 2020). Similarly, we identified 9–11 TMDs in the putative poplar Lsi2 homologs except Potri.017G141950.1, which contained five TMDs (Supplementary Fig. 1A). Only Potri.012G144000.5 contained a signal peptide (Supplementary Fig. 1), further supporting the notion that this gene is the putative poplar Lsi2 homolog corresponding to OsLsi2. A recent study (Nawaz et al. 2020) identified a highly conserved region with a consensus sequence “XXTKHXWFXXCXXXXRX” among the Lsi2 proteins in different plant species such as rice, wheat, and Sorghum. This conserved sequence, however, was not found in the Lsi2 homologs in poplar. Many plants with high Si accumulation capacity, such as Equisetum (horsetail) and pumpkin, do not contain this conserved sequence either (Mitani-Ueno et al. 2011a; Grégoire et al. 2012). Therefore, this conserved sequence might not have any functional importance, but further characterization is warranted. Lsi2 is not an extensively characterized protein like Lsi1, and thus there is a little information available for this protein.
Siliplant 1 (Slp1) protein
Our genome-wide analysis identified 27 proteins in the studied plant species which belong to the Slp1 protein family. Out of the 27 proteins, there were only two poplar proteins which will be referred to as PtSlp1a (Potri.004G168600.10) and PtSlp1b (Potri.009G129900.6) in the remaining text. The isoelectric point (pI value) of these proteins was computed as positively charged AAs are involved in biosilicification (Otzen 2012; Nawaz et al. 2020; Kumar et al. 2020). The predicted pI values of the poplar Slp1 homologs were 9.96 and 9.78 (Table 1) for PtSlp1a and Ptslp1b, respectively. Similar to the known Slp1 protein in Sorghum, PtSlp1a and PtSlp1b are rich in proline (23.5% in PtSlp1a and 32.2% in PtSlp1b) and lysine (13.7% in PtSlp1a and 15.4% in PtSlp1b) (Table 1). However, they are poor in histidine (3.7% in PtSlp1a and 2.1% in PtSlp1b), whereas the known Sorghum Slp1 protein is rich in histidine (Kumar et al. 2020). We also tested whether these proteins have signal peptides because Sorghum Slp1 is a secretory protein (Kumar et al. 2020). Both poplar Slp1 homologs contain a signal peptide (Supplementary Fig. 1B).
The reported peptide sequence for in vitro silica precipitation is HKKPVPPKPKPEPK (Kumar et al. 2020). In addition, the well-characterized Sorghum Slp1 protein has three conserved domains, namely the A-, B- and C-domains. The A-domain is rich in histidine and aspartic acid (i.e., H, D- rich) and the consensus sequence is DXFHKXHDYHXXXXHFH, whereas the B- and C- domains are rich in proline, lysine, glutamic acid (P, K, E- rich) and proline, threonine, and tyrosine (P, T, Y-rich) with consensus sequences KKPXPXKPKPXPKPXPXPX and YHXPXPTYXSPTPIYHPPX, respectively (Kumar et al. 2020). We looked for the presence of these features in the putative Slp1 poplar homologs. We found that both PtSlp1a and PtSlp1b contain the peptide sequences reported for in vitro silica precipitation, but these sequences are partially conserved. The peptide is also more conserved in PtSlp1a than PtSlp1b. When screening for the conserved A-, B-, and C- domains, we found that PtSlp1a and PtSlp1b do not contain H, D-rich regions (i.e., A-domain). They do contain the P, K, E-rich and P, T, Y-rich regions which are partially conserved relative to the Sorghum Slp1 protein (Fig. 5A). Absence of the H, D-rich domain suggests that PtSlp1a or PtSlp1b might be a moderate Si accumulator as described by (Kumar et al. 2020). Finally, we checked the PtSlp1a/PtSlp1b protein sequences for the presence of any repeat AA sequences as the Sorghum Slp1 contains multiple repeats (Kauss et al. 2003; Kumar et al. 2020). We found that PtSlp1a contains a sequence (KPKPPVFKPPPVPI) repeated six times, whereas PtSlp1b harbors a sequence (PKPKPPIFK) which is repeated three times (Fig. 5B). The AA lengths of PtSlp1a and PtSlp1b are also different. PtSlp1a has 513 AA whereas the length of PtSlp1b is 336 AA. The length of the Sorghum Slp1 protein is 524 AA which is almost equivalent to PtSlp1a. These results suggest that PtSlp1a is the best candidate Slp1 homolog in poplar.
Screening Slp1 signature sequence in the putative poplar homologs. A Alignment of two identified poplar Slp1 homologs along with the known Slp1 protein of Sorghum. The signature domains of Slp1 proteins are indicated with colored boxes (A-domain is in pink, B-domain is in sky blue, C-domain is in red). B Repeat sequences found in the two poplar Slp1 homolog proteins identified in this study PtSlp1a (Potri.004G168600.10) and PtSlp1b (Potri.009G129900.6)
Gene expression pattern of Si transporters in poplar
In plants with high Si accumulation capabilities, channel-type SIT genes (i.e., Lsi1/Lsi6) and Lsi2 are predominantly expressed in root and stem tissue (Jadhao et al. 2020), consistent with their role of Si uptake by roots, whereas Slp1 is expressed in the inflorescence and leaves (Kumar et al. 2020). We examined the expression pattern of the putative channel-type SITs, Lsi2, and Slp1 homologs in poplar using gene expression data from the ePlant poplar (http://bar.utoronto.ca/eplant_poplar/) database. We found that the expression levels of both PtLsi1 (Potri.017G083300) and PtLsi2 (Potri.012G144000) were lower in the root than in the other parts of the plant (Fig. 6A), such as leaves, suggesting that poplar is a weak Si accumulator. PtSlp1a (Potri.004G168600) is highly expressed in the inflorescence and young leaves (Fig. 6A), similar to the Sorghum Slp1 gene (Nawaz et al. 2020; Kumar et al. 2020). Furthermore, the expression of PtSlp1a was found to be much higher than that of PtSlp1b (Potri.009G129900) in multiple tissue types (Fig. 6A). This expression pattern suggests that PtSlp1a might be the best candidate Slp1 homolog in poplar, which is consistent with our inference based on the analysis of protein sequences in section “Siliplant 1 (Slp1) protein”. Similar to observations above, analysis of expression patterns across the P. trichocarpa GWAS population also revealed that expression levels of these four genes was highest in leaves followed by roots and was negligible in xylem tissues (Fig. 6B). Moreover, there was significant variation across genotypes for each gene. When evaluated across the P. trichocarpa tissue Atlas expression profiles, Potri.004G168600 consistently had the highest expression across conditions followed by Potri.009G129900. Across all conditions, Potri.017G083300 consistently showed low expression levels (Fig. 6C).
Gene expression profiles of poplar Si transporters (PtLsi1; Potri.017G083300 and PtLsi2; Potri.012G144000) and the candidate silicification proteins (PtSlp1a; Potri.004G168600 and PtSlp1b; Potri.009G129900). A Digital expression patterns obtained from gene expression data available at http://bar.utoronto.ca/eplant_poplar/. B Expression profiles of different Si transporters across the P. trichocarpa GWAS population. C Expression profiles of Si transporters across different tissue types
Potential interaction partners of PtLsi1 and PtLsi2
We identified the putative interaction partners of PtLsi1/PtLsi2 using public data in the STRING database (Szklarczyk et al. 2021). The putative interaction partners of PtLsi1 (Potri.017G083300) include different types of ion transporters, such as POPTR_0013s05030.2 (Fig. 7A), which is an aquaporin protein. This protein belongs to the major intrinsic protein (MIP) family and small basic intrinsic protein (SIP) subfamily and is involved in the transport of water and ions into the cell (Johanson and Gustavsson 2002; Ishikawa et al. 2005). Other important interaction partners of PtLsi1 are POPTR _0005s20790.1 and POPTR_0002s07550.2, which are involved in a range of metabolic pathways. The interaction partners of PtLsi2 include Delta-1-pyrroline-5-carboxylate synthase (POPTR_0008 s06060.1, POPTR_0010s20590.1), aminomethyltransferase (gdcT1/gdcT2), and the 26S proteasome regulatory subunit family protein. Among these interaction partners, Delta-1-pyrroline-5-carboxylate synthase (P5Cs) plays a key role in proline biosynthesis in plants which helps protect the plant from different biotic and abiotic stressors (Hayat et al. 2012; Dar et al. 2016). This suggests that PtLsi2 may play a role in stress response.
Predicted interaction partners of the putative Si transporters in poplar. A PtLsi1 (Potri.017G083300; UniProt accession# B9IJY1) and B PtLsi2 (Potri.012G144000; UniProt accession# U5FXI4). The network was obtained from the STRING database (Szklarczyk et al. 2021)
Evolution of the Slp1 gene family
Gene duplication plays an important role in plant evolution. Specifically, gene family expansion and genomic evolutionary mechanisms mainly depend on gene duplication events. In this study, gene duplication events were identified in the Slp1 gene family using a bioinformatics analysis. All identified Slp1 genes were mapped to their corresponding chromosomes, as shown in Fig. 8. Some of these genes showed tandem gene duplications such as Sobic.001G265900 versus Sobic.001G266100 in Sorghum, Sobic.001G438400 versus Sobic.001G438500 in Sorghum, and LOC_Os03g14130 versus LOC_Os03g14140 on rice chromosome 3 (Fig. 8). The expansion of the Slp1 gene family partially resulted from segmental duplications, such as a segmental duplication on Sorghum chromosome 1 (e.g., the segment containing Sobic.001G265900 and Sobic.001G266100 versus the segment containing Sobic.001G438400 and Sobic.001G438500) as well as a segmental duplication between the two poplar chromosomes (the segment containing Potri.004G168600 versus the segment containing Potri.009G129900) (Fig. 8). There were clear syntenic genomic blocks conserved among different plant species, such as the block containing LOC_Os03g14130 and LOC_Os03g14140 in rice and the block containing Sobic.001G438400 and Sobic.001G438500 in Sorghum. These results indicate that the evolution and expansion of the Slp1 gene family was mediated by tandem gene duplication and segmental duplications within the same chromosome and between different chromosomes.
Mutation profiles of Si transporters across the P. trichocarpa GWAS population
Among the poplar Si transporters identified in this study, Potri.004G168600 had the highest number of mutations with three significant mutations based on SNPeff analysis. This gene had a codon deletion mutation at position Chr04:18772192, a non-synonymous mutation (G > A) at position Chr04:18772384, and another non-synonymous mutation (T > C) at position Chr04:18772686. Potri.009G129900 had the highest impact mutation at position Chr09:10591818 where a cytosine insertion created a frame-shift mutation (GTT > GCTT). The frame-shift mutation occurred only in the heterozygous condition in 103 of the 917 P. trichocarpa genotypes evaluated. The absence of the homozygous state of this allele might suggest deleterious effects if protein function is significantly affected. The second-most impactful mutation occurred in Potri.012G144000 at position Chr12:15654656 with a four-nucleotide deletion from the reference allele creating an alternative splice site (CATATATA > CATA). Of the 917 genotypes, 450 carried the CATA allele in the homozygous state while 366 were homozygous and 101 individuals carried the CATATATA minor allele in a homozygous state. The relative lack of high-impact mutations in these genes might be indicative of their essential role in P. trichocarpa biology. Genotypes exhibiting high-impact mutations identified from this study can be further characterized to evaluate the putative impacts on trait expression.
Protein 3D structure analysis
Figure 9 shows the 3D structures of the putative Si transporter and silicification proteins of poplar. The 3D structures were predicted with Alphafold and the accuracy of these structures was determined by the predicted local distance difference test (pLDDT) score. pLDDT scores range from 0 to 100 and a higher value indicates a more accurate prediction. Another indicator for assessing the Alphafold predicted protein structure is the predicted aligned error (PAE) which assesses the confidence in the domain packing and large-scale topology of the protein. PAE is presented as an interactive 2D plot where dark blue indicates small error and red indicates large error. Alphafold detected a well-defined 3D structure (Fig. 9A) for most of the PtLsi1 with high confidence (pLDDT > 80; Fig. 9H) except the C-terminus (residue 1–32) which was depicted as a ribbon-like structure with a low pLDDT score (pLDDT < 50; Fig. 9G). This ribbon-like structure is likely to be an intrinsically disordered region (IDR). The PtLsi1 protein also exhibited high structural similarity with OsLsi1 except the IDR which is detected in the N-terminus in OsLsi1. Disordered protein domains are often important for intracellular signaling and can transition from a disordered to an ordered state upon binding to other proteins (Ruff and Pappu 2021; David et al. 2022). Therefore, the presence of an IDR in a different location in PtLsi1 versus OsLsi1 indicates that these two proteins may have different binding partners or function differently. PtLsi2 also showed a well-folded structure predicted by Alphafold and was highly similar to the OsLsi2 3D structure (Fig. 9C, D) with a pLDDT score > 80 (Fig. 9G). Interestingly, the IDR detected in both proteins were found in very similar positions (residues 211–267 in PtLsi2 and 211–295 in OsLsi2). The high structural similarity along with a similar location of the IDR implies that PtLsi2 is the candidate homolog for OsLsi2 in poplar. For silicification proteins, both the candidates in poplar (PtSlp1a and PtSlp1b) and the known SbSlp1 in Sorghum showed highly disordered structures in the Alphafold-predicted 3D model (Fig. 9E–G) with pLDDT scores < 50 (Fig. 9G). Comparing PtSlp1a and PtSlp1b with SbSlp1, it appears that PtSlp1a is structurally more similar to SbSlp1 indicating that PtSlp1a may be the candidate silicification protein in poplar.
3D structures of the Si transporter and silicification proteins in poplar and rice. A–G Alphafold predicted 3D structures with the interactive 2D plots of predicted aligned error (PAE). Dark blue in the PAE plot indicates smaller errors whereas red indicates larger errors. H Interactive 2D plot of predicted local difference distance (pLDDT) scores for different models. The pLDDT plot gives the best information on intra-domain confidence with a value > 90 indicative of a high score
The quality and accuracy of the Alphafold prediction model varies. The pLDDT score is a good guide for model quality and it correlates well with model accuracy when assessed against an experimentally determined structure. However, further experimental characterization and structure-based mutational studies are required to validate the structural model. Another limitation of Alphafold is that the predictions yield a static picture of protein structure and contain little or no information on protein dynamics. Conformational changes in proteins are often key to their function and regulation and are frequently triggered by binding to other molecules (e.g., proteins or small molecules/ligands) or by post-translational modifications. The current prediction algorithm is unable to model these conformational changes and is therefore largely unable to provide a crucial aspect of understanding the protein structure–function relationship (Jumper et al. 2021; van Breugel et al. 2022).
Conclusion
Previous studies on Si transporters mainly focused on monocot plants, particularly food crops. In this study, we identified transporter genes putatively involved in Si uptake, transport, and deposition in the woody bioenergy crop, P. trichocarpa. Further, we identified putative interaction partners and expression profiles of Si transporter genes and have described the possible evolution of Si transporter proteins in P. trichocarpa. To the best of our knowledge, this study represents the first attempt to provide an in-depth analysis of Si transporters in P. trichocarpa. The current findings expand our understanding of Si transporters in woody plants and provide knowledge that will greatly enhance genetic engineering strategies aimed at improving carbon sequestration and biomass quality of bioenergy feedstocks. Specifically, different Si transporter genes can be combined and expressed together in P. trichocarpa to examine their effects on phytolith formation. An additional next step would examine if the expression of rice Lsi6, the homolog that was not identified in poplar, improves phytolith formation in P. trichocarpa.
References
Ahanger MA, Bhat JA, Siddiqui MH et al (2020) Integration of silicon and secondary metabolites in plants: a significant association in stress tolerance. J Exp Bot 71:6758–6774. https://doi.org/10.1093/jxb/eraa291
Almagro Armenteros JJ, Sønderby CK, Sønderby SK et al (2017) DeepLoc: prediction of protein subcellular localization using deep learning. Bioinformatics 33:3387–3395. https://doi.org/10.1093/bioinformatics/btx431
Armenteros JJA, Tsirigos KD, Sønderby CK et al (2019) SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat Biotechnol 37:420–423. https://doi.org/10.1038/s41587-019-0036-z
Bailey TL, Williams N, Misleh C, Li WW (2006) MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res 34:W369–W373. https://doi.org/10.1093/nar/gkl198
Bateman A, Birney E, Cerruti L et al (2002) The Pfam protein families database. Nucleic Acids Res 30:276–280. https://doi.org/10.1093/nar/30.1.276
Buchfink B, Xie C, Huson DH (2015) Fast and sensitive protein alignment using DIAMOND. Nat Methods 12:59–60. https://doi.org/10.1038/nmeth.3176
Chiba Y, Mitani N, Yamaji N, Ma JF (2009) HvLsi1 is a silicon influx transporter in barley. Plant J 57:810–818. https://doi.org/10.1111/j.1365-313X.2008.03728.x
Dar MI, Naikoo MI, Rehman F et al (2016) Proline accumulation in plants: roles in stress tolerance and plant development. In: Iqbal N, Nazar R, Nafees A, Khan N (eds) Osmolytes and plants acclimation to changing environment: emerging omics technologies. Springer, New Delhi, pp 155–166
David A, Islam S, Tankhilevich E, Sternberg MJE (2022) The alphafold database of protein structures: a biologist’s guide. J Mol Biol 434:167336. https://doi.org/10.1016/j.jmb.2021.167336
Deshmukh RK, Vivancos J, Guérin V et al (2013) Identification and functional characterization of silicon transporters in soybean using comparative genomics of major intrinsic proteins in Arabidopsis and rice. Plant Mol Biol 83:303–315. https://doi.org/10.1007/s11103-013-0087-3
Deshmukh RK, Vivancos J, Ramakrishnan G et al (2015) A precise spacing between the NPA domains of aquaporins is essential for silicon permeability in plants. Plant J 83:489–500. https://doi.org/10.1111/tpj.12904
Enright AJ, Van Dongen S, Ouzounis CA (2002) An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res 30:1575–1584. https://doi.org/10.1093/nar/30.7.1575
Gasteiger E, Hoogland C, Gattiker A et al (2005) Protein identification and analysis tools on the expasy server. In: Walker JM (ed) The proteomics protocols handbook. Humana Press, Totowa, pp 571–607
Gaur S, Kumar J, Kumar D et al (2020) Fascinating impact of silicon and silicon transporters in plants: a review. Ecotoxicol Environ Saf 202:110885. https://doi.org/10.1016/j.ecoenv.2020.110885
Goodstein DM, Shu S, Howson R et al (2012) Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res 40:D1178–D1186. https://doi.org/10.1093/nar/gkr944
Grégoire C, Rémus-Borel W, Vivancos J et al (2012) Discovery of a multigene family of aquaporin silicon transporters in the primitive plant Equisetum arvense. Plant J 72:320–330. https://doi.org/10.1111/j.1365-313X.2012.05082.x
Hans Wedepohl K (1995) The composition of the continental crust. Geochim Cosmochim Acta 59:1217–1232. https://doi.org/10.1016/0016-7037(95)00038-2
Hayat S, Hayat Q, Alyemeni MN et al (2012) Role of proline under changing environments: a review. Plant Signal Behav 7:1456–1466. https://doi.org/10.4161/psb.21949
Horton P, Park K-J, Obayashi T et al (2007) WoLF PSORT: protein localization predictor. Nucleic Acids Res 35:W585–W587. https://doi.org/10.1093/nar/gkm259
Ishikawa F, Suga S, Uemura T et al (2005) Novel type aquaporin SIPs are mainly localized to the ER membrane and show cell-specific expression in Arabidopsis thaliana. FEBS Lett 579:5814–5820. https://doi.org/10.1016/j.febslet.2005.09.076
Jadhao KR, Bansal A, Rout GR (2020) Silicon amendment induces synergistic plant defense mechanism against pink stem borer (Sesamia inferens Walker) in finger millet (Eleusine coracana Gaertn). Sci Rep 10:4229
Johanson U, Gustavsson S (2002) A new subfamily of major intrinsic proteins in plants. Mol Biol Evol 19:456–461. https://doi.org/10.1093/oxfordjournals.molbev.a004101
Jones P, Binns D, Chang H-Y et al (2014) InterProScan 5: genome-scale protein function classification. Bioinformatics 30:1236–1240. https://doi.org/10.1093/bioinformatics/btu031
Jumper J, Evans R, Pritzel A et al (2021) Highly accurate protein structure prediction with AlphaFold. Nature 596:583–589. https://doi.org/10.1038/s41586-021-03819-2
Kanehisa M, Sato Y, Morishima K (2016) Blastkoala and ghostkoala: KEGG tools for functional characterization of genome and metagenome sequences. J Mol Biol 428:726–731. https://doi.org/10.1016/j.jmb.2015.11.006
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. https://doi.org/10.1093/molbev/mst010
Kaur H, Greger M (2019) A review on si uptake and transport system. Plants. https://doi.org/10.3390/plants8040081
Kauss H, Seehaus K, Franke R et al (2003) Silica deposition by a strongly cationic proline-rich protein from systemically resistant cucumber plants. Plant J 33:87–95. https://doi.org/10.1046/j.1365-313x.2003.01606.x
Krogh A, Larsson B, von Heijne G, Sonnhammer ELL (2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305:567–580. https://doi.org/10.1006/jmbi.2000.4315
Kulmanov M, Hoehndorf R (2020) DeepGOPlus: improved protein function prediction from sequence. Bioinformatics 36:422–429. https://doi.org/10.1093/bioinformatics/btz595
Kumar S, Soukup M, Elbaum R (2017) Silicification in grasses: variation between different cell types. Front Plant Sci 8:438. https://doi.org/10.3389/fpls.2017.00438
Kumar S, Stecher G, Li M et al (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549. https://doi.org/10.1093/molbev/msy096
Kumar S, Adiram-Filiba N, Blum S et al (2020) Siliplant1 protein precipitates silica in sorghum silica cells. J Exp Bot 71:6830–6843. https://doi.org/10.1093/jxb/eraa258
Ma JF, Yamaji N (2015) A cooperative system of silicon transport in plants. Trends Plant Sci 20:435–442. https://doi.org/10.1016/j.tplants.2015.04.007
Ma JF, Tamai K, Yamaji N et al (2006) A silicon transporter in rice. Nature 440:688–691. https://doi.org/10.1038/nature04590
Ma JF, Yamaji N, Mitani N et al (2007) An efflux transporter of silicon in rice. Nature 448:209–212. https://doi.org/10.1038/nature05964
Ma JF, Yamaji N, Mitani-Ueno N (2011) Transport of silicon from roots to panicles in plants. Proc Jpn Acad Ser B 87:377–385. https://doi.org/10.2183/pjab.87.377
Madeira F, Park YM, Lee J et al (2019) The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res 47:W636–W641. https://doi.org/10.1093/nar/gkz268
Minh BQ, Schmidt HA, Chernomor O et al (2020) IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol 37:1530–1534. https://doi.org/10.1093/molbev/msaa015
Mirdita M, Schütze K, Moriwaki Y et al (2022) ColabFold: making protein folding accessible to all. Nat Methods 19:679–682. https://doi.org/10.1038/s41592-022-01488-1
Mistry J, Chuguransky S, Williams L et al (2021) Pfam: the protein families database in 2021. Nucleic Acids Res 49:D412–D419. https://doi.org/10.1093/nar/gkaa913
Mitani N, Chiba Y, Yamaji N, Ma JF (2009a) Identification and characterization of maize and barley Lsi2-like silicon efflux transporters reveals a distinct silicon uptake system from that in rice. Plant Cell 21:2133–2142. https://doi.org/10.1105/tpc.109.067884
Mitani N, Yamaji N, Ma JF (2009b) Identification of maize silicon influx transporters. Plant Cell Physiol 50:5–12. https://doi.org/10.1093/pcp/pcn110
Mitani N, Yamaji N, Ago Y et al (2011) Isolation and functional characterization of an influx silicon transporter in two pumpkin cultivars contrasting in silicon accumulation. Plant J 66:231–240. https://doi.org/10.1111/j.1365-313X.2011.04483.x
Mitani-Ueno N, Yamaji N, Ma JF (2011a) Silicon efflux transporters isolated from two pumpkin cultivars contrasting in Si uptake. Plant Signal Behav 6:991–994. https://doi.org/10.4161/psb.6.7.15462
Mitani-Ueno N, Yamaji N, Zhao F-J, Ma JF (2011b) The aromatic/arginine selectivity filter of NIP aquaporins plays a critical role in substrate selectivity for silicon, boron, and arsenic. J Exp Bot 62:4391–4398. https://doi.org/10.1093/jxb/err158
Montpetit J, Vivancos J, Mitani-Ueno N et al (2012) Cloning, functional characterization and heterologous expression of TaLsi1, a wheat silicon transporter gene. Plant Mol Biol 79:35–46. https://doi.org/10.1007/s11103-012-9892-3
Nawaz MA, Azeem F, Zakharenko AM et al (2020) In-silico exploration of channel type and efflux silicon transporters and silicification proteins in 80 sequenced viridiplantae genomes. Plants. https://doi.org/10.3390/plants9111612
Omasits U, Ahrens CH, Müller S, Wollscheid B (2014) Protter: interactive protein feature visualization and integration with experimental proteomic data. Bioinformatics 30:884–886. https://doi.org/10.1093/bioinformatics/btt607
Otzen D (2012) The role of proteins in biosilicification. Scientifica (cairo) 2012:867562. https://doi.org/10.6064/2012/867562
Ruff KM, Pappu RV (2021) Alphafold and implications for intrinsically disordered proteins. J Mol Biol 433:167208. https://doi.org/10.1016/j.jmb.2021.167208
Sahu SS, Loaiza CD, Kaundal R (2020) Plant-mSubP: a computational framework for the prediction of single- and multi-target protein subcellular localization using integrated machine-learning approaches. AoB Plants. https://doi.org/10.1093/aobpla/plz068
Sakurai G, Satake A, Yamaji N et al (2015) In silico simulation modeling reveals the importance of the Casparian strip for efficient silicon uptake in rice roots. Plant Cell Physiol 56:631–639. https://doi.org/10.1093/pcp/pcv017
Sakurai G, Yamaji N, Mitani-Ueno N et al (2017) A model of silicon dynamics in rice: an analysis of the investment efficiency of si transporters. Front Plant Sci 8:1187. https://doi.org/10.3389/fpls.2017.01187
Souri Z, Khanna K, Karimi N, Ahmad P (2020) Silicon and plants: current knowledge and future prospects. J Plant Growth Regul. https://doi.org/10.1007/s00344-020-10172-7
Sun H, Guo J, Duan Y et al (2017) Isolation and functional characterization of CsLsi1, a silicon transporter gene in Cucumis sativus. Physiol Plant 159:201–214. https://doi.org/10.1111/ppl.12515
Szklarczyk D, Gable AL, Nastou KC et al (2021) The STRING database in 2021: customizable protein–protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res 49:D605–D612. https://doi.org/10.1093/nar/gkaa1074
Van Bel M, Diels T, Vancaester E et al (2018) PLAZA 4.0: an integrative resource for functional, evolutionary and comparative plant genomics. Nucleic Acids Res 46:D1190–D1196. https://doi.org/10.1093/nar/gkx1002
van Breugel M, Silva RE, Andreeva A (2022) Structural validation and assessment of AlphaFold2 predictions for centrosomal and centriolar proteins and their complexes. Commun Biol 5:312. https://doi.org/10.1038/s42003-022-03269-0
Vatansever R, Ozyigit II, Filiz E, Gozukirmizi N (2017) Genome-wide exploration of silicon (Si) transporter genes, Lsi1 and Lsi2 in plants; insights into Si-accumulation status/capacity of plants. Biometals 30:185–200. https://doi.org/10.1007/s10534-017-9992-2
Vivancos J, Deshmukh R, Grégoire C et al (2016) Identification and characterization of silicon efflux transporters in horsetail (Equisetum arvense). J Plant Physiol 200:82–89. https://doi.org/10.1016/j.jplph.2016.06.011
Waese J, Fan J, Pasha A et al (2017) ePlant: visualizing and exploring multiple levels of data for hypothesis generation in plant biology. Plant Cell 29:1806–1821. https://doi.org/10.1105/tpc.17.00073
Wallace IS, Roberts DM (2004) Homology modeling of representative subfamilies of arabidopsis major intrinsic proteins. classification based on the aromatic/arginine selectivity filter. Plant Physiol 135:1059–1068. https://doi.org/10.1104/pp.103.033415
Wang Y, Tang H, Debarry JD et al (2012) MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res 40:e49. https://doi.org/10.1093/nar/gkr1293
Waterhouse AM, Procter JB, Martin DMA et al (2009) Jalview version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25:1189–1191. https://doi.org/10.1093/bioinformatics/btp033
Yamaji N, Mitatni N, Ma JF (2008) A transporter regulating silicon distribution in rice shoots. Plant Cell 20:1381–1389. https://doi.org/10.1105/tpc.108.059311
Yang X, Hu R, Yin H et al (2017) The Kalanchoë genome provides insights into convergent evolution and building blocks of crassulacean acid metabolism. Nat Commun 8:1899. https://doi.org/10.1038/s41467-017-01491-7
Yu C-S, Chen Y-C, Lu C-H, Hwang J-K (2006) Prediction of protein subcellular localization. Proteins 64:643–651. https://doi.org/10.1002/prot.21018
Zhang J, Yang Y, Zheng K et al (2018) Genome-wide association studies and expression-based quantitative trait loci analyses reveal roles of HCT2 in caffeoylquinic acid biosynthesis and its regulation by defense-responsive transcription factors in Populus. New Phytol 220:502–516. https://doi.org/10.1111/nph.15297
Funding
This work was supported by the Laboratory Directed Research and Development program of Oak Ridge National Laboratory. This manuscript has been authored by UT-Battelle, LLC under Contract No. DE-AC05-00OR22725 with the U.S. Department of Energy. The United States Government retains and the publisher, by accepting the article for publication, acknowledges that the United States Government retains a non-exclusive, paid-up, irrevocable, worldwide license to publish or reproduce the published form of this manuscript, or allow others to do so, for United States Government purposes. The Department of Energy will provide public access to these results of federally sponsored research in accordance with the DOE Public Access Plan (http://energy.gov/downloads/doe-public-access-plan).
Author information
Authors and Affiliations
Contributions
MMH, XY and DW conceived the idea. MMH, XY, WM, KF and TBY analyzed the data. MMH, SM, WM and XY led the writing and revision of the manuscript. DW, SM, NAG, GY, MZM contributed to the manuscript revision. All authors accepted the final version of the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
11816_2022_788_MOESM1_ESM.pptx
Supplementary file1 Transmembrane domains (TMD) in the putative Lsi2 (A) and Slp1 (B) homologs in poplar. Red regions indicate a signal peptide (PPTX 2350 KB)
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Hassan, M.M., Martin, S., Feng, K. et al. Genome-wide identification and functional prediction of silicon (Si) transporters in poplar (Populus trichocarpa). Plant Biotechnol Rep 17, 285–302 (2023). https://doi.org/10.1007/s11816-022-00788-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11816-022-00788-4