Abstract
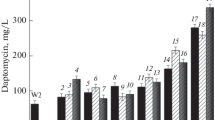
To improve the yield of daptomycin by Streptomyces roseosporus, a method of rational screening of He–Ne Laser and N-methyl-N-nitro-N-nitrosoguanidine (NTG)-induced mutants was employed in this work. Under the optimal mutagenesis conditions (NTG of 0.3 mg/L, 40 min; irradiation at 15 mW, 15 min), two steps of combined mutations were conducted. Screening of mutants was done according to the individual resistance to n-decanoic acid and daptomycin. A mutant strain LC-54-16, with the highest daptomycin production ability of 616 mg/L was obtained, which was over 5 times higher than the wild strain LC-52-6. The transcription levels of the main dpt genes in the mutant were approximately five times higher than those in the wild. The superiority of the mutant to the wild strain was further proved by the comparative studies on the kinetics of the mutant and the wild. The decrease of the inhibition of substrate and product was further confirmed, as well. It was concluded that the method of rational screening of He–Ne Laser and NTG-induced mutants could efficiently improve the daptomycin production ability of S. roseosporus.







Similar content being viewed by others
Abbreviations
- Xmax (gL−1):
-
Maximum cell dry weight
- X0 (gL−1):
-
Initial cell dry weight
- μmax (h−1):
-
Maximum specific growth rate
- K S (gL−1):
-
Saturation coefficient of glucose
- Pmax (g L−1):
-
Maximum daptomycin titer
- α:
-
Growth associated product formation rate constant
- β (h−1):
-
Non-growth-associated product formation rate constant
- YX/S (gg−1):
-
Cell yield coefficient of glucose
- YP/S (gg−1):
-
Product yield coefficient of glucose
- m (gg−1):
-
Cell maintenance coefficient
- Smax (gL−1):
-
Maximum glucose concentration at which specific growth rate is zero
- n S :
-
Exponent of glucose repression
- \( S_{de}^{{ \max }}\left( {{\hbox{g}}{{\hbox{L}}^{ - {1}}}} \right) \) :
-
Maximum n-decanoic acid concentration at which specific growth rate is zero
- n de :
-
Exponent of n-decanoic acid repression
- \( S_{da}^{{ \max }}\left( {{\hbox{g}}{{\hbox{L}}^{ - {1}}}} \right) \) :
-
Maximum n-decanoic acid concentration at which specific growth rate is zero
- n da :
-
Exponent of daptomycin repression
- \( {Y_{P/{S_{de}}}} \) :
-
Product yield coefficient of n-decanoic acid
- X (gL−1):
-
Biomass concentration
- S (gL−1):
-
Glucose concentration
- S de (gL−1):
-
n-decanoic acid concentration
- P (gL−1):
-
Daptomycin concentration
References
Debono, M., Barnhart, M., Carrell, C. B., Hoffmann, J. A., Occolowitz, J. L., Abbott, B. J., et al. (1987). The Journal of Antibiotics, 40, 761–777.
Huber, F. M., Pieper, R. L., & Tietz, A. J. (1988). Journal of Biotechnology, 7, 283–292.
Zhou, P. F., Wang, Y. W., Guo, M., Wang, H. H., & Xie, J. P. (2009). Review of Medical Microbiology, 20, 12–18.
Chamberlain, R. S., Culshaw, D. L., Donovan, B. J., & Lamp, K. C. (2009). Surgery, 14, 6316–6324.
Arbeit, R. D., Maki, D., Tally, F. P., Campanaro, E., & Eisenstein, B. I. (2004). Clinical Infectious Diseases, 38, 1673–1681.
Kirkpatrick, P., Raja, A., LaBonte, J., & Lebbos, J. (2003). Nature Reviews. Drug Discovery, 2, 943–944.
Miao, V., Coeffet-LeGal, M. F., Brian, P., Brost, R., Penn, J., Whiting, A., et al. (2005). Microbiology-Sgm, 151, 1507–1523.
Yeats, C., Bentley, S., & Bateman, A. (2003). BMC. Microbiology. 2003. Available from: http://www.biomedcentral.com/1471-2180/3/3. Accessed February 6, 2003.
Nguyen, K. T., Kau, D., Gu, J. Q., Brian, P., Wrigley, S. K., Baltz, R. H., et al. (2006). Molecular Microbiology, 61, 1294–1307.
Olano, C., Lombo, F., Mendez, C., & Salas, J. A. (2008). Metabolic Engineering, 10, 281–292.
Khaliq, S., Akhtar, K., Ghauri, M. A., Iqbal, R., Khalid, A. M., & Muddassar, M. (2009). Microbiological Research, 164, 469–477.
Wu, Z. F., Bajpai, R. K., & Yan, W. (2008). Biocatalysis and Biotransformation, 26, 444–449.
Kohli, R., Bose, B., & Gupta, P. K. (2001). Journal of Photochemistry and Photobiology. B: Biology, 60, 136–142.
Kumar, K. S., Manimaran, A., Permaul, K., & Singh, S. J. (2009). Bioscience and Bioengineering, 107, 494–498.
Rhee, K. H., & Julian, D. (2006). Journal of Microbiology and Biotechnology, 16, 1841–1848.
Livak, K. J., & Schmittgen, T. D. (2001). Methods, 25, 402–408.
Coeffet-Le Gal, M. F., Thurston, L., Rich, P., Miao, V., & Baltz, R. H. (2006). Microbiology-Sgm, 152, 2993–3001.
Luong, J. H. T. (1987). Biotechnology and Bioengineering, 29, 242–248.
Maiti, S. K., Singh, K. P., Lantz, A. E., Bhushan, M., & Wangikar, P. P. (2010). Biotechnology and Bioengineering, 105, 109–120.
Fouchard, S., Pruvost, J., Degrenne, B., Titica, M., & Legrand, J. (2009). Biotechnology and Bioengineering, 102, 232–245.
Dogan, G. (2007). System Dynamics Review, 23, 415–436.
Acknowledgments
The authors wish to acknowledge the financial support provided by the National High Technology Research, Development Program of China (2007AA02Z200), the National Natural Science Foundation of China (No. 20706042, No. 20906070), and the Program of Introducing Talents of Discipline to Universities (No. B06006).
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Supplementary Table 1
Primers used for RT-PCR (DOC 42 kb)
Rights and permissions
About this article
Cite this article
Yu, G., Jia, X., Wen, J. et al. Strain Improvement of Streptomyces roseosporus for Daptomycin Production by Rational Screening of He–Ne Laser and NTG Induced Mutants and Kinetic Modeling. Appl Biochem Biotechnol 163, 729–743 (2011). https://doi.org/10.1007/s12010-010-9078-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-010-9078-x