Abstract
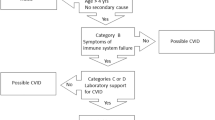
Common variable immunodeficiency disorders (CVID) are an enigmatic group of often heritable conditions, which may manifest for the first time in early childhood or as late as the eighth decade of life. In the last 5 years, next generation sequencing (NGS) has revolutionised identification of genetic disorders. However, despite the best efforts of researchers around the globe, CVID conditions have been slow to yield their molecular secrets. We have previously described the many clinical advantages of identifying the genetic basis of primary immunodeficiency disorders (PIDs). In a minority of CVID patients, monogenic defects have now been identified. If a causative mutation is identified, these conditions are reclassified as CVID-like disorders. Here we discuss recent advances in the genetics of CVID and discuss how NGS can be optimally deployed to identify the causal mutations responsible for the protean clinical manifestations of these conditions. Diagnostic criteria such as the Ameratunga et al. criteria will continue to play an important role in patient management as well as case selection and sequencing strategy design until the genetic conundrum of CVID is solved.

Similar content being viewed by others
References
Gathmann B, Mahlaoui N, Gerard L et al (2014) Clinical picture and treatment of 2212 patients with common variable immunodeficiency. J Allergy Clin Immunol 134(1):116–126
Chapel H, Lucas M, Lee M et al (2008) Common variable immunodeficiency disorders: division into distinct clinical phenotypes. Blood 112(2):277–286
Ameratunga R, Becroft DM, Hunter W (2000) The simultaneous presentation of sarcoidosis and common variable immune deficiency. Pathology 32(4):280–282
Ameratunga R, Woon ST, Gillis D, Koopmans W, Steele R (2013) New diagnostic criteria for common variable immune deficiency (CVID), which may assist with decisions to treat with intravenous or subcutaneous immunoglobulin. Clin Exp Immunol 174(2):203–211
Ameratunga R, Woon ST, Gillis D, Koopmans W, Steele R (2014) New diagnostic criteria for CVID. Expert Rev Clin Immunol 10(2):183–186
Koopmans W, Woon ST, Zeng IS et al (2013) Variability of memory B cell markers in a cohort of common variable immune deficiency patients over six months. Scand J Immunol 77(6):470–475
Ameratunga R, Barker R, Steele R et al (2015) Profound reversible hypogammaglobulinemia caused by celiac disease in the absence of protein losing enteropathy. J Clin Immunol 35(6):589–594
Ameratunga R, Lindsay K, Woon S-T, Jordan A, Anderson NE, Koopmans W (2015) New diagnostic criteria could distinguish common variable immunodeficiency disorder from anticonvulsant-induced hypogammaglobulinemia. Clin Exp Neuroimmunol 6(1):83–88
Ameratunga R, Storey P, Barker R, Jordan A, Koopmans W, Woon ST (2015) Application of diagnostic and treatment criteria for common variable immunodeficiency disorder. Expert Rev Clin Immunol 12(3):257–266
Ameratunga RV, Parry S, Kenedi C (2013) Hypogammaglobulinemia factitia- Munchausen syndrome presenting as common variable immune deficiency. Allergy Asthma Clin Immunol. 9(36)
Ameratunga R, Brewerton M, Slade C et al (2014) Comparison of diagnostic criteria for common variable immunodeficiency disorder. Front Immunol 5:415
Bonilla FA, Barlan I, Chapel H et al (2016) International Consensus Document (ICON): common variable immunodeficiency disorders. J Allergy Clin Immunol Pract 4(1):38–59
Oksenhendler E, Gerard L, Fieschi C et al (2008) Infections in 252 patients with common variable immunodeficiency. Clin Infect Dis 46(10):1547–1554
Conley ME, Notarangelo LD, Etzioni A (1999) Diagnostic criteria for primary immunodeficiencies. Representing PAGID (Pan-American Group for Immunodeficiency) and ESID (European Society for Immunodeficiencies). Clin Immunol 93(3):190–197
Empson M, Sinclair J, O'Donnell J, Ameratunga R, Fitzharris P, Steele R (2004) The assessment and management of primary antibody deficiency. N Z Med J 117(1195):U914
Ameratunga R, Gillis D, Steele R (2016) Diagnostic criteria for common variable immunodeficiency disorders. J Allergy Clin Immunol Pract 4(5):1017–1018
Richards S, Aziz N, Bale S et al (2015) Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med: Off J Am Coll Med Genet 17(5):405–424
Woon ST, Ameratunga R. Comprehensive genetic testing for primary immunodeficiency disorders in a tertiary hospital: 10-year experience in Auckland, New Zealand. Allergy Asthma Clin Immunol 2016;12:65
Ameratunga R, Winkelstein JA, Brody L et al (1998) Molecular analysis of the third component of canine complement (C3) and identification of the mutation responsible for hereditary canine C3 deficiency. J Immunol 160(6):2824–2830
Bacchelli C, Buckland KF, Buckridge S et al (2011) The C76R transmembrane activator and calcium modulator cyclophilin ligand interactor mutation disrupts antibody production and B-cell homeostasis in heterozygous and homozygous mice. J Allergy Clin Immunol 127(5):1253–1259 e1213
Ameratunga R, Bartlett A, McCall J, Steele R, Woon ST, Katelaris CH (2016) Hereditary angioedema as a metabolic liver disorder: novel therapeutic options and prospects for cure. Front Immunol 7:547
Fliegauf ML, Bryant V, Frede N et al (2015) Haploinsufficiency of the NF-κB1 subunit p50 in common variable immunodeficiency. Am J Hum Genet 97(3):389–403
Bogaert DJ, Dullaers M, Lambrecht BN, Vermaelen KY, De Baere E, Haerynck F (2016) Genes associated with common variable immunodeficiency: one diagnosis to rule them all? J Med Genet. 53(9):575–590. https://doi.org/10.1136/jmedgenet-2015-103690
Raje N, Soden S, Swanson D, Ciaccio CE, Kingsmore SF, Dinwiddie DL (2014) Utility of next generation sequencing in clinical primary immunodeficiencies. Curr Allergy Asthma Rep 14(10):468
Grimbacher B, Warnatz K, Yong PF, Korganow AS, Peter HH (2016) The crossroads of autoimmunity and immunodeficiency: lessons from polygenic traits and monogenic defects. J Allergy Clin Immunol 137(1):3–17 quiz 18
Ameratunga R, Woon ST, Brewerton M et al (2011) Primary immune deficiency disorders in the South Pacific: the clinical utility of a customized genetic testing program in New Zealand. Ann N Y Acad Sci 1238:53–64
Ameratunga R, Woon ST, Neas K, Love DR (2010) The clinical utility of molecular diagnostic testing for primary immune deficiency disorders: a case based review. Allergy Asthma Clin Immunol 6(1):12
Ameratunga R, Steele R, Jordan A et al (2016) The case for a national service for primary immune deficiency disorders in New Zealand. N Z Med J. 129(1436):75–90
Ameratunga R, Woon ST (2009) Customised molecular diagnosis of primary immune deficiency disorders in New Zealand: an efficient strategy for a small developed country. N Z Med J 122(1304):46–53
Rodriguez-Cortez VC, Del Pino-Molina L, Rodriguez-Ubreva J et al (2015) Monozygotic twins discordant for common variable immunodeficiency reveal impaired DNA demethylation during naive-to-memory B-cell transition. Nat Commun 6:7335
van Schouwenburg PA, Davenport EE, Kienzler AK et al (2015) Application of whole genome and RNA sequencing to investigate the genomic landscape of common variable immunodeficiency disorders. Clin Immunol 160(2):301–314
Kienzler AK, Hargreaves CE, Patel SY (2017) The role of genomics in common variable immunodeficiency disorders. Clin Exp Immunol 188(3):326–332
Ameratunga R, Koopmans W, Woon ST, et al. Epistatic interactions between mutations of TACI (TNFRS13B) and TCF3 result in a severe primary immunodeficiency disorder and systemic lupus erythematosus. Clin Transl Immunol (Nature). 2017;(in press)
Bateson (1909) Discussion on the influence of heredity on disease, with special reference to tuberculosis, cancer, and diseases of the nervous system: introductory address. Proc R Soc Med;2(Gen Rep):22–30
Koopmans W, Woon ST, Brooks AE, Dunbar PR, Browett P, Ameratunga R (2013) Clinical variability of family members with the C104R mutation in transmembrane activator and calcium modulator and cyclophilin ligand interactor (TACI). J Clin Immunol 33(1):68–73
Acknowledgements
We are extremely grateful to our patients for allowing us to undertake these studies for the benefit of others. We hope this discovery will benefit them and any future families with these disorders. We thank AMRF, A+ Trust, IDFNZ, ASCIA, and the Australian National Health and Medical Research Council (NHMRC, Program Grant 1054925) and Project 1127198 for grant support. We also receive support from Bloody Long Way (BLW). C.S. is supported by NHMRC postgraduate scholarship 1075666. We thank Dr. Kitty Croxson, ADHB, and LabPlus management for ongoing support. Bioinformatic analysis was supported by the New Zealand eScience Infrastructure. All studies were approved by Auckland Hospital (3435), NZ Ministry of Health (MEC/06/10/134), and the Walter and Eliza Hall Institute (WEHI) Human Research Ethics Committee (HREC 10/02).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
All authors declare they have no conflicts of interest.
Authorship
RA- wrote the first draft.
KL- wrote bioinformatics section.
STW, DG, and RS edited and revised the manuscript.
CS and VB contributed to the text.
Rights and permissions
About this article
Cite this article
Ameratunga, R., Lehnert, K., Woon, ST. et al. Review: Diagnosing Common Variable Immunodeficiency Disorder in the Era of Genome Sequencing. Clinic Rev Allerg Immunol 54, 261–268 (2018). https://doi.org/10.1007/s12016-017-8645-0
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12016-017-8645-0