Abstract
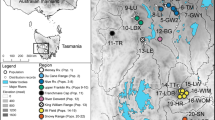
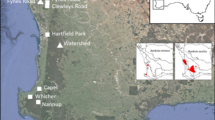
Reliance on clonal reproduction is associated with an increased risk of extinction. Grevillea infecunda is a rare, putatively sterile shrub restricted to 11 populations in a localized coastal region of south-eastern Australia. We assessed the genetic diversity, clonal diversity and spatial distribution of clones in all populations of G. infecunda to guide conservation management. Eight chloroplast haplotypes were identified from the trnL – trnF and trnQ – rps16 intergenic spacer regions. All individuals belonged to a single maternal lineage dominated by one haplotype (95/111 samples). Minor haplotypes differed from the common haplotype only by single mutational steps. However, microsatellite markers revealed 89 multilocus genotypes (MLGs) in 38 multilocus lineages (MLLs) of variable size. MLLs were not shared among populations and ramets from different MLLs rarely intermingled physically. New shoots arising after fire were confirmed to belong to previously-existing MLLs, indicating that this species exhibits adaptation to fire. Genetically similar MLGs were more likely to be found in close proximity than less similar MLGs, resulting in significant spatial autocorrelation to ca 350 m. Genetic diversity was moderate but genotypic diversity was low once likely clonality was taken into account. Clonality appears to have arisen several times within the holly-leafed grevilleas and examining G. infecunda is a step towards understanding why and how often clonality occurs, and the long-term evolutionary outcomes of this life history.




Similar content being viewed by others
References
Ally D, Ritland K, Otto SP (2008) Can clone size serve as a proxy for clone age? An exploration using microsatellite divergence in Populus tremuloides. Molec Ecol 17:4897–4911
Ally D, Ritland K, Otto SP (2010) Aging in a long-lived clonal tree. PLoS Biol 8: e1000454
Andrews S FastQC. Available at http://www.bioinformatics.bbsrc.ac.uk/projects/fastqc
Arnaud-Haond S, Duarte CM, Alberto F, Serrao EA (2007) Standardizing methods to address clonality in population studies. Mol Ecol 16:5115–5139
Balloux F, Lehmann L, de Meeus, T (2003) The population genetics of clonal and partially clonal diploids. Genetics 164:1635–1644
Barrett SC (2015) Influences of clonality on plant sexual reproduction. PNAS 112:8859–8866
Boers NM, Haig R, Schnute J (2004) PBS Mapping 2: developer's guide. Canadian Technical Report of Fisheries and Aquatic Sciences 2550
Bonin A, Bellemain E, Bronken Eidesen P, Pompanon F, Brochmann C, Taberlet,P (2004) How to track and assess genotyping errors in population genetics studies. Molec Ecol 13:3261–3273
Bragg L M, Stone G, Butler MK, Hugenholtz P, Tyson GW (2013) Shining a light on dark sequencing: characterising errors in Ion Torrent PGM data. PLoS Computat Biol 9:e1003031
Bretagnolle F, Thompson JD (1993) Tansley Review No. 78. Gametes with the somatic chromosome number: mechanisms of their formation and role in the evolution of autopolyploid plants. New Phytol 129:1–22
Burne HM, Yates CJ, Ladd PG (2003) Comparative population structure and reproductive biology of the critically endangered shrub Grevillea althoferorum and two closely related more common congeners. Biol Conservation 114 53–65
Caddy HAR, Gross CL (2006) Population structure and fecundity in the putative sterile Shrub, Grevillea rhizomatosa Olde and Marriott (Proteaceae). Proc Linn Soc New South Wales 127:11–18
Cadotte MW, Cavender-Bares J, Tilman D, Oakley TH (2009) Using phylogenetic, functional and trait diversity to understand patterns of plant community productivity. PLoS One 4:e5695
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Gonzalez Pena A, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R (2010) QIIME allows analysis of high-throughput community sequencing data. Nature, Meth 7:335–336
Carter O (2006) National recovery plan for the Anglesea Grevillea Grevillea infecunda. Dept Sustain and Env, Melbourne
Clark LV, Jasieniuk M (2011) POLYSAT: an R package for polyploidy microsatellite analysis. Molec Ecol Resources 11:562–566
Clement M, Posada D, Crandall KA (2000) TCS: a computer program to estimate gene genealogies. Molec Ecol 9:1657–1660
de Witte L C, Stöcklin J (2010) Longevity of clonal plants: why it matters and how to measure it. Ann Bot (Oxford) 106:859–870
Dorken ME, Eckert CG (2001) Severely reduced sexual reproduction in northern populations of a clonal plant, Decodon verticillatus (Lythraceae). J Ecol 89:339–350
Douhovnikoff V, Dodd RS (2003) Intra-clonal variation and a similarity threshold for identification of clones: application to Salix exigua using AFLP molecular markers. Theor Appl Genet 106:1307–1315
Dunlop M, Hilbert D, Ferrier S, House A, Liedloff A, Prober SM, Smyth A, Martin TG, Harwood T, Williams KJ, Fletcher C, Murphy H (2012) The implications of climate change for biodiversity conservation and the National Reserve System: Final synthesis. A report prepared for the Department of Sustainability, Environment, Water, Population and Communities, and the Department of Climate Change and Energy Efficiency. CSIRO Climate Adaptation Flagship, Canberra
Eckert CG (2002) The loss of sex in clonal plants. Evol Ecol 15:501–520
England PR, Ayre DJ, Whelan RJ (1999) Microsatellites in the Australian shrub Grevillea macleayana (Proteaceae). Molec Ecol 8:685–702
Frankham R, Ballou JD, Briscoe DA (2010) Introduction to Conservation Genetics. Cambridge, UK, Cambridge University Press
Gitzendanner MA, Weekley CW, Germain-Aubrey CC, Soltis DE, Soltis PS (2011) Microsatellite evidence for high clonality and limited genetic diversity in Ziziphus celata (Rhamnaceae), an endangered, self-incompatible shrub endemic to the Lake Wales Ridge, Florida, USA. Conservation Genet 13:223–234
Gross CL, Caddy HAR (2006) Are differences in breeding mechanisms and fertility among populations contributing to rarity in Grevillea rhizomatosa (Proteaceae)? Amer J Bot 93:1791–1799
Gross CL, Nelson PA, Haddadchi A, Fatemi M (2012) Somatic mutations contribute to genotypic diversity in sterile and fertile populations of the threatened shrub, Grevillea rhizomatosa (Proteaceae). Ann Bot (Oxford) 109:331–342
Hoebee SE (2011) Development and cross-species amplification of microsatellite markers from the endangered Wee Jasper Grevillea (Grevillea iaspicula, Proteaceae). Muelleria 29:93–96
Holmes GD, Downing TL, James EA, Blacket MJ, Hoffmann AA, Bayly MJ. 2014. Phylogeny of the holly grevilleas based on nuclear ribosomal and chloroplast DNA. Austral Syst Bot 27:55–77
Holmes GD, James EA, Hoffmann AA, (2009). Divergent levels of genetic variation and ploidy among populations of the rare shrub, Grevillea repens (Proteaceae). Conservation Genet 10:827–837
Honnay O, Bossuyt B (2005) Prolonged clonal growth: escape route or route to extinction? Oikos 108:427–432
Honnay O, Jacquemyn, J (2007) Susceptibility of common and rare plant species to the genetic consequences of habitat fragmentation. Conservation Biol 21: 823–31
James EA, Brown GK, Citroen R, Blacket MJ (2012) Microsatellite marker development for two species of holly-leafed Grevillea and cross-species amplification in the Aspleniifolia/Hookeriana Subgroup (Proteaceae). Conservation Genet Resources 4:137–140
James EA, McDougall KL (2014) Spatial genetic structure reflects extensive clonality, low genotypic diversity and habitat fragmentation in Grevillea renwickiana (Proteaceae), a rare, sterile shrub from south-eastern Australia. Ann Bot (Oxford) 114:413–423
Jombart T (2008) adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 24:1403–1405
Jombart T, Devillard S, Dufour AB, Pontier D (2008) Revealing cryptic spatial patterns in genetic variability by a new multivariate method. Heredity 101:92–103
Jombart T (2011). A tutorial for the spatial Analysis of Principal Components (sPCA) using adegenet 1.3-0. Available at: cran.r-project.org/web/packages/adegenet/vignettes/adegenet-spca.pdf
Jombart T (2012). An introduction to adegenet 1.3-5. Downloaded from http://adegenet.r-forge.r-project.org/
Joyce B (2004) The young volcanic regions of Southeastern Australia: early studies, physical volcanology, and eruption risk. Proc Roy Soc Victoria 116:1–13
Kimpton SK, James EA, Drinnan AN (2002) Reproductive biology and genetic marker diversity in Grevillea infecunda (Proteaceae), a rare plant with no known seed production. Austral Syst Bot 15:485–492
Klekowski EJ (2003) Plant clonality, mutation, diplontic selection and mutational meltdown. Biol J Linn Soc 79:61–67
Lacey CJ (1974) Rhizomes in tropical eucalypts and their role in recovery from fire damage. Austral J Bot 22:29–38
Lo EY, Stefanovic S, Ritland K, Dickinson TA (2010) Fine-scale comparisons of genetic variability in seed families of asexually and sexually reproducing Crataegus (hawthorn; Rosaceae). Amer J Bot 97:1014–1024
Lohse M, Bolger AM, Nagel A, Fernie AR, Lunn JE, Stitt M, and e. al. 2012. RobiNA: a user-friendly, integrated software solution for RNA-Seq-based transcriptomics. Nucleic Acids Res 40:W622-W627
Lynch AJJ, Barnes RW, Cambecedes J, Vaillancourt RE (1998) Genetic evidence that Lomatia tasmanica (Proeaceae) is an ancient clone. Austral J Bot 46:25–33
Makinson RO (2000) Grevillea. ABRS/CSIRO, Melbourne, Australia
Maynard Smith J (1978) The evolution of sex. Cambridge University Press, Cambridge, UK
McGillivray DJ (1986) New names in Grevillea (Proteaceae) 7. Castle Hill: New South Wales, DJ McGillivray 16p.-. En Many sp. nov. Geog= 0 Systematics: ANGIOSPERMAE (PROTEACEAE: GREVILLEA) (KR, 198603012)
McGillivray DJ, Makinson RO (1993) Grevillea; Proteaceae. A taxonomic revision. Melbourne University Press, Carlton
Meirmans PG, Van Tienderen PH (2004) genotype and genodive: two programs for the analysis of genetic diversity of asexual organisms. Molec Ecol Notes 4:792–794
Millar A, Byrne M, Coates DJ (2010) The maintenance of disparate levels of clonality, genetic diversity and genetic differentiation in disjunct subspecies of the rare Banksia ionthocarpa. Molec Ecol 19:4217–4227
Mock KE, Rowe CA, Hooten MB, Dewoody J, Hipkins VD (2008) Clonal dynamics in western North American aspen (Populus tremuloides). Molec Ecol 17:4827–4844
Novaes RM, De Lemos Filho JP, Ribeiro RA, Lovato MB (2010) Phylogeography of Plathymenia reticulata (Leguminosae) reveals patterns of recent range expansion towards northeastern Brazil and southern Cerrados in Eastern Tropical South America. Molec Ecol 19:985–998
Olde P, Marriott N (1995) The Grevillea Book Volume 2. Kangaroo Press Ltd, Kenthurst
Otto SP, Whitton J (2000) Polyploid incidence and evolution. Annual Rev Genet 34:401–437
Peakall R, Smouse PE (2012) GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research – an update. Bioinformatics 28:2537–2539
Peter BM, Slatkin M (2013) Detecting range expansions from genetic data. Evolution 67:3274–3289
Piquot Y, Valero M, Cuguen J, de Laguerie P, Vernet P (1998) Variation in sexual and asexual reproduction among young and old populations of the perennial macrophyte Sparganium erectum. Oikos 82:139–148
Pompanon F, Bonin A, Bellemain E, Taberlet P (2005) Genotyping errors: causes, consequences and solutions. Nat Rev Genet 6:847–859
Quinlan AR, Hall IM (2010) BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26:841–842
R Core Team (2014) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. Available at: http://www.R-project.org
Richards A J (1997) Plant Breeding Systems. Chapman and Hall, London
Schaal, BA, Leverich WJ (1996) Molecular variation in isolated plant populations. Pl Spec Biol 11:33–40
Silvertown JW (2008) The evolutionary maintenance of sexual reproduction: evidence from the ecological distribution of asexual reproduction in clonal plants. Int J Pl Sci 169:157–168
Silvertown, JW, Doust JL (1993) Introduction to plant population biology. Blackwell, Oxford, UK
Smith JA (2004) Reproductive, developmental and genetic factors associated with sterility in the endangered, clonal shrub Hakea pulvinifera L.A.S. Johnson (Proteaceae). Dissertation, University of New England.
Suzuki J-I, Herben T, Maki M (2005) An under-appreciated difficulty: sampling of plant populations for analysis using molecular markers. Evol Ecol 18:625–646
Thuiller W, Albert C, Araújo MB, Berry PM, Cabeza M, Guisan A, Hickler T, Midgley GF, Paterson J, Schurr FM, Sykes MT, Zimmermann NE (2008) Predicting global change impacts on plant species’ distributions: Future challenges. Perspect Pl Ecol, Evol Syst 9:137–152
Vallejo-Marín M, Dorken ME, Barrett SCH (2010) The Ecological and Evolutionary Consequences of Clonality for Plant Mating. Annual Rev Ecol Evol Syst 41:193–213
van Kleunen M, Fischer M, Schmid B (2001) Effects of intraspecific competition on size variation and reproductive allocation in a clonal plant. Oikos 94:515–524
van Kleunen M, Fischer M, Schmid B (2002) Experimental life-history evolution: Selection on the allocation to sexual reproduction and its plasticity in a clonal plant. Evolution 56:2168–2177
Whitton J, Sears CJ, Baack EJ, Otto SP (2008) The Dynamic Nature of Apomixis in the Angiosperms. Int J Pl Sci 169:169–182
Acknowledgements
We are grateful to Alcoa Australia for financial support. Cybec Foundation provided funding for Marc Freestone who was the recipient of a Willis Summer Studentship and contributed to the project. Alisdair Macleod (Australian Automotive Research Centre, Anglesea, VIC) provided access to population PG. We also thank Margaret McDonald, ANGAIR and members of the G. infecunda Recovery Team for much appreciated advice, and Gavan McCarthy for assistance in the field.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest. This study was carried out under DSE Scientific Permit 10006581 and complies with permit requirements and ethical standards.
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 2495 kb)
Rights and permissions
About this article
Cite this article
James, E.A., Jordan, R., Brown, G.K. et al. Assessing the genetic legacy of a rare, clonal Australian shrub Grevillea infecunda (Proteaceae). Folia Geobot 52, 387–400 (2017). https://doi.org/10.1007/s12224-016-9258-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12224-016-9258-8