Abstract
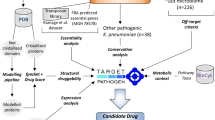
The emergence and dissemination of pan drug resistant clones of Klebsiella pneumoniae are great threat to public health. In this regard new therapeutic targets must be highlighted to pave the path for novel drug discovery and development. Subtractive proteomic pipeline brought forth UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase (LpxC), a Zn+2 dependent cytoplasmic metalloprotein and catalyze the rate limiting deacetylation step of lipid A biosynthesis pathway. Primary sequence analysis followed by 3-dimensional (3-D) structure elucidation of the protein led to the detection of K. pneumoniae LpxC (KpLpxC) topology distinct from its orthologous counterparts in other bacterial species. Molecular docking study of the protein recognized receptor antagonist compound 106, a uridine-based LpxC inhibitory compound, as a ligand best able to fit the binding pocket with a Gold Score of 67.53. Molecular dynamics simulation of docked KpLpxC revealed an alternate binding pattern of ligand in the active site. The ligand tail exhibited preferred binding to the domain I residues as opposed to the substrate binding hydrophobic channel of subdomain II, usually targeted by inhibitory compounds. Comparison with the undocked KpLpxC system demonstrated ligand induced high conformational changes in the hydrophobic channel of subdomain II in KpLpxC. Hence, ligand exerted its inhibitory potential by rendering the channel unstable for substrate binding.











Similar content being viewed by others
References
Paczosa MK, Mecsas J (2016) Klebsiella pneumoniae: going on the offense with a strong defense. Microbiol Mol Biol Rev 80:629–661
Band VI, Satola SW, Burd EM, Farley MM, Jacob JT, Weiss DS (2018) Carbapenem-resistant Klebsiella pneumoniae exhibiting clinically undetected colistin heteroresistance leads to treatment failure in a murine model of infection. mBio 9:02448-17
Farhadi T, Fakharian A, Ovchinnikov RS (2017) Virtual screening for potential inhibitors of CTX-M-15 protein of Klebsiella pneumoniae. Interdiscip Sci Comput Life Sci. https://doi.org/10.1007/s12539-017-0222-y
Nordmann P, Cuzon G, Naas T (2009) The real threat of Klebsiella pneumoniae carbapenemase-producing bacteria. Lancet Infect Dis 9:228–236
Center for Disease Control and Prevention (CDC) (2013) Antibiotic resistance threats in the United States. U.S Department of Health and Human Services
Mantzarlis K, Makris D, Manoulakas E, Karvouniaris M, Zakynthinos E (2013) Risk factors for the first episode of Klebsiella pneumoniae resistant to carbapenems infection in critically ill patients: a prospective study. BioMed Res Int 2013:850547. https://doi.org/10.1155/2013/850547
Tzouvelekis LS, Markogiannakis A, Psichogiou M, Tassios PT, Daikos GL (2012) Carbapenemases in Klebsiella pneumoniae and other Enterobacteriaceae: an evolving crisis of global dimensions. Clin Microbiol Rev 25:682–707
Phoebe Chen YP, Chen F (2008) Identifying targets for drug discovery using bioinformatics. Expert Opin Ther Target 12:383–389
Searls DB (2000) Using bioinformatics in gene and drug discovery. Drug Discov Today 5:135–143
Sliwoski G, Kothiwale S, Meiler J, Lowe EW (2014) Computational methods in drug discovery. Pharmacol Rev 66:334–395
Ahmad S, Raza S, Uddin R, Azam SS (2017) Binding mode analysis, dynamic simulation and binding free energy calculations of the MurF ligase from Acinetobacter baumannii. J Mol Graph Model 77:72–85
Butt AM, Nasrullah I, Tahir S, Tong Y (2012) Comparative genomics analysis of Mycobacterium ulcerans for the identification of putative essential genes and therapeutic candidates. PLoS One 7:e43080
Chawley P, Samal HB, Prava J, Suar M, Mahapatra RK (2014) Comparative genomics study for identification of drug and vaccine targets in Vibrio cholerae: MurA ligase as a case study. Genomics 103:83–93
Singh S, Singh DB, Singh A, Gautam B, Ram G, Dwivedi S, Ramteke PW (2016) An approach for identification of novel drug targets in Streptococcus pyogenes SF370 through pathway analysis. Interdiscip Sci Comput Life Sci 8:388–394
Vijayalakshmi P, Nisha J, Rajalakshmi M (2014) Virtual screening of potential inhibitor against FtsZ protein from Staphylococcus aureus. Interdiscip Sci Comput Life Sci 6:331–339
Hosen MI, Tanmoy AM, Mahbuba DA, Salma U, Nazim M, Islam MT, Akhteruzzaman S (2014) Application of a subtractive genomics approach for in silico identification and characterization of novel drug targets in Mycobacterium tuberculosis F11. Interdiscip Sci Comput Life Sci 6:48–56
Uddin R, Siddiqui QN, Azam SS, Saima B, Wadood A (2018) Identification and characterization of potential druggable targets among hypothetical proteins of extensively drug resistant Mycobacterium tuberculosis (XDR KZN 605) through subtractive genomics approach. Eur J Pharm Sci 114:13–23
Wadood A, Jamal A, Riaz M, Khan A, Uddin R, Jelani M, Azam SS (2017) Subtractive genome analysis for in silico identification and characterization of novel drug targets in Streptococcus pneumonia strain JJA. Microb Pathog 115:194–198
Azam SS, Abbasi SW, Akhtar AS, Mirza ML (2014) Comparative modeling and molecular docking studies of d-alanine: d-alanine ligase: a target of antibacterial drugs. Med Chem Res 23:4108–4137
Azam SS, Abro A, Raza S, Saroosh A (2014) Structure and dynamics studies of sterol 24-C-methyltransferase with mechanism based inactivators for the disruption of ergosterol biosynthesis. Mol Biol Rep 41:4279–4293
Pérez-Castillo Y, Froeyen M, Cabrera-Pérez M, Nowé A (2011) Molecular dynamics and docking simulations as a proof of high flexibility in E. coli FabH and its relevance for accurate inhibitor modeling. J Comput Aided Mol Des 25:371–393
Sinko W, de Oliveira C, Williams S, Van Wynsberghe A, Durrant JD, Cao R, Oldfield E, McCammon JA (2011) Applying molecular dynamics simulations to identify rarely sampled ligand-bound conformational states of undecaprenyl pyrophosphate synthase, an antibacterial target. Chem Biol Drug Des 77:412–420
Barb AW, Zhou P (2008) Mechanism and inhibition of LpxC: an essential zinc-dependent deacetylase of bacterial lipid A synthesis. Curr Pharm Biotechnol 9:9–15
Yethon JA, Whitfield C (2001) Lipopolysaccharide as a target for the development of novel therapeutics in gram-negative bacteria. Curr Drug Targets Infect Disord 1:91–106
Mdluli KE, Witte PR, Kline T, Barb AW, Erwin AL, Mansfield BE, McClerren AL, Pirrung MC, Tumey LN, Warrener P, Raetz CR (2006) Molecular validation of LpxC as an antibacterial drug target in Pseudomonas aeruginosa. Antimicrob Agents Chemother 50:2178–2184
Magrane M, UniProt C (2011) UniProt knowledgebase: a hub of integrated protein data. Database (oxford). https://doi.org/10.1093/database/bar009 (bar 009)
Boutet E, Lieberherr D, Tognolli M, Schneider M, Bairoch A (2007) UniProtKB/Swiss Prot. Methods Mol Biol 406:89–112
Huang Y, Niu B, Gao Y, Fu L, Li W (2010) CD-HIT Suite: a web server for clustering and comparing biological sequences. Bioinformatics 26:680–682
Li W, Godzik A (2006) Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22:1658–1659
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Pruitt KD, Tatusova T, Maglott DR (2007) NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res 35:61–65
Zhang R, Ou HY, Zhang CT (2004) DEG: a database of essential genes. Nucleic Acids Res 32:271–272
Zhang R, Lin Y (2009) DEG 5.0, a database of essential genes in both prokaryotes and eukaryotes. Nucleic Acids Res 37:D455-D458
Moriya Y, Itoh M, Okuda S, Yoshizawa AC, Kanehisa M (2007) KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res 35:182–185
Kanehisa M, Goto S (2000) KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 28:27–30
Wishart DS, Knox C, Guo AC, Cheng D, Shrivastava S, Tzur D, Gautam B, Hassanali M (2008) DrugBank: a knowledgebase for drugs, drug actions and drug targets. Nucleic Acids Res 36:D901-D906
Yu CS, Chen YC, Lu CH, Hwang JK (2006) Prediction of protein subcellular localization. Proteins Struct Funct Bioinform 64:643–651
Nancy YY, Wagner JR, Laird MR, Melli G, Rey S, Lo R, Dao P, Sahinalp SC, Ester M, Foster LJ, Brinkman FS (2010) PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 26:1608–1615
Fiser A (2010) Template-based protein structure modeling. Methods Mol Biol 673:73–94
Šali, Blundell TL (1993) Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol 234:779–815
Zhang Y (2008) I-TASSER server for protein 3D structure prediction. BMC Bioinform 9:40
Pieper U, Webb BM, Dong GQ, Schneidman-Duhovny D, Fan H, Kim SJ, Khuri N, Spill YG, Weinkam P, Hammel M, Tainer JA (2014) ModBase, a database of annotated comparative protein structure models and associated resources. Nucleic Acids Res 42:336–346
Bates PA, Kelley LA, MacCallum RM, Sternberg MJ (2001) Enhancement of protein modeling by human intervention in applying the automatic programs 3D-JIGSAW and 3D-PSSM. Proteins 45:39–46
Arnold K, Bordoli L, Kopp J, Schwede T (2006) The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22:195–201
Lambert C, Leonard N, De Bolle X, Depiereux E (2002) ESyPred3D: prediction of proteins 3D structures. Bioinformatics 18:1250–1256
Laskowski RA, MacArthur MW, Moss DS, Thornton JM (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Cryst 26:283–291
Colovos C, Yeates TO (1993) Verification of protein structures: patterns of nonbonded atomic interactions. Protein Sci 2:1511–1519
Eisenberg D, Lüthy R, Bowie JU (1997) VERIFY3D: Assessment of protein models with three-dimensional profiles. Methods Enzymol 277:396–404
Wiederstein M, Sippl MJ (2007) ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res 35:407–410
Meng EC, Pettersen EF, Couch GS, Huang CC, Ferrin TE (2006) Tools for integrated sequence-structure analysis with UCSF Chimera. BMC Bioinform 7:339
Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE (2004) UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612
Cole KE, Gattis SG, Angell HD, Fierke CA, Christianson DW (2010) Structure of the metal-dependent deacetylase LpxC from Yersinia enterocolitica complexed with the potent inhibitor CHIR-090. Biochemistry 50:258–265
Mochalkin I, Knafels JD, Lightle S (2008) Crystal structure of LpxC from Pseudomonas aeruginosa complexed with the potent BB-78485 inhibitor. Protein Sci 17:450–457
Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, Thompson JD (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7:539
Schomburg I, Chang A, Placzek S, Söhngen C, Rother M, Lang M, Munaretto C, Ulas S, Stelzer M, Grote A, Scheer M (2013) BRENDA in 2013: integrated reactions, kinetic data, enzyme function data, improved disease classification: new options and contents in BRENDA. Nucleic Acids Res 41:764–772
Barb AW, Leavy TM, Robins LI, Guan Z, Six DA, Zhou P, Bertozzi CR, Raetz CR (2009) Uridine-based inhibitors as new leads for antibiotics targeting Escherichia coli LpxC. Biochemistry 48:3068–3077
Clements JM, Coignard F, Johnson I, Chandler S, Palan S, Waller A, Wijkmans J, Hunter MG (2002) Antibacterial activities and characterization of novel inhibitors of LpxC. Antimicrob Agents Chemother 46:1793–1799
Coggins BE, McClerren AL, Jiang L, Li X, Rudolph J, Hindsgaul O, Raetz CR, Zhou P (2005) Refined solution structure of the LpxC–TU-514 complex and pK a analysis of an active site histidine: insights into the mechanism and inhibitor design. Biochemistry 44:1114–1126
Gennadios HA, Whittington DA, Li X, Fierke CA, Christianson DW (2006) Mechanistic inferences from the binding of ligands to LpxC, a metal-dependent deacetylase. Biochemistry 45:7940–7948
Jackman JE, Raetz CR, Fierke CA (2001) Site-directed mutagenesis of the bacterial metalloamidase UDP-(3-O-acyl)-N-acetylglucosamine deacetylase (LpxC). Identification of the zinc binding site. Biochemistry 40:514–523
Kadam RU, Garg D, Chavan A, Roy N (2007) Evaluation of Pseudomonas aeruginosa deacetylase lpxc inhibitory activity of Dual PDE4–TNFα inhibitors: a multiscreening approach. J Chem Inf Model 47:1188–1195
Kadam RU, Garg D, Roy N (2008) Selective mapping of chemical space for Pseudomonas aeruginosa deacetylase LpxC inhibitory potential. Chem Biol Drug Des 71:45–56
Kline T, Andersen NH, Harwood EA, Bowman J, Malanda A, Endsley S, Erwin AL, Doyle M, Fong S, Harris AL, Mendelsohn B (2002) Potent, novel in vitro inhibitors of the Pseudomonas aeruginosa deacetylase LpxC. J Med Chem 45:3112–3129
Pirrung MC, Tumey LN, Raetz CR, Jackman JE, Snehalatha K, McClerren AL, Fierke CA, Gantt SL, Rusche KM (2002) Inhibition of the antibacterial target UDP-(3-O-acyl)-N-acetylglucosamine deacetylase (LpxC): isoxazoline zinc amidase inhibitors bearing diverse metal binding groups. J Med Chem 45:4359–4370
Pirrung MC, Tumey LN, McClerren AL, Raetz CR (2003) High-throughput catch-and-release synthesis of oxazoline hydroxamates. Structure–activity relationships in novel inhibitors of Escherichia coli LpxC: in vitro enzyme inhibition and antibacterial properties. J Am Chem Soc 125:1575–1586
Mansoor UF, Vitharana D, Reddy PA, Daubaras DL, McNicholas P, Orth P, Black T, Siddiqui MA (2011) Design and synthesis of potent Gram-negative specific LpxC inhibitors. Bioorg Med Chem Lett 21:1155–1161
Brown MF, Reilly U, Abramite JA, Arcari JT, Oliver R, Barham RA, Che Y, Chen JM, Collantes EM, Chung SW, Desbonnet C (2012) Potent inhibitors of LpxC for the treatment of Gram-negative infections. J Med Chem 55:914–923
McAllister LA, Montgomery JI, Abramite JA, Reilly U, Brown MF, Chen JM, Barham RA, Che Y, Chung SW, Menard CA, Mitton-Fry M (2012) Heterocyclic methylsulfone hydroxamic acid LpxC inhibitors as Gram-negative antibacterial agents. Bioorg Med Chem Lett 22:6832–6838
Montgomery JI, Brown MF, Reilly U, Price LM, Abramite JA, Arcari J, Barham R, Che Y, Chen JM, Chung SW, Collantes EM (2012) Pyridone methylsulfone hydroxamate LpxC inhibitors for the treatment of serious gram-negative infections. J Med Chem 55:1662–1670
Warmus JS, Quinn CL, Taylor C, Murphy ST, Johnson TA, Limberakis C, Ortwine D, Bronstein J, Pagano P, Knafels JD, Lightle S (2012) Structure based design of an in vivo active hydroxamic acid inhibitor of P. aeruginosa LpxC. Bioorg Med Chem Lett 22:2536–2543
Li Z, Wan H, Shi Y, Ouyang P (2004) Personal experience with four kinds of chemical structure drawing software: review on ChemDraw, ChemWindow, ISIS/Draw, and ChemSketch. J Chem Inf Comput Sci 44:1886–1890
Jones G, Willett P, Glen RC (1995) Molecular recognition of receptor sites using a genetic algorithm with a description of desolvation. J Mol Biol 245:43–53
Trott O, Olson AJ (2010) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 31:455–461
Humphrey W, Dalke A, Schulten K (1996) VMD: visual molecular dynamics. J Mol Graph 14:33–38
Wallace AC, Laskowski RA, Thornton JM (1995) LIGPLOT a program to generate schematic diagrams of protein-ligand interactions. Protein Eng 8:127–134
Accelrys Software Inc (2012) Discovery studio visualizer, Release 3.5. Accelrys Software Inc., San Diego
Weiner PK, Kollman PA (1981) AMBER: assisted model building with energy refinement. A general program for modeling molecules and their interactions. J Comput Chem 2:287–303
Yang W, Riley BT, Lei X, Porebski BT, Kass I, Buckle AM, McGowan S (2017) Generation of AMBER force field parameters for zinc centres of M1 and M17 family aminopeptidases. J Biomol Struct Dyn 28:1–10
Dickerson JE, Zhu A, Robertson DL, Hentges KE (2011) Defining the role of essential genes in human disease. PLoS One 6:e27368
Galperin MY, Koonin EV (1999) Searching for drug targets in microbial genomes. Curr Opin Biotechnol 10:571–578
Sakharkar KR, Sakharkar MK, Chow VT (2004) A novel genomics approach for the identification of drug targets in pathogens, with special reference to Pseudomonas aeruginosa. In Silico Biol 4:355–360
Bakheet TM, Doig AJ (2010) Properties and identification of antibiotic drug targets. BMC Bioinform 11:195
Fauman EB, Rai BK, Huang ES (2011) Structure-based druggability assessment—identifying suitable targets for small molecule therapeutics. Curr Opin Chem Biol 15:463–468
Hale MR, Hill P, Lahiri S, Miller MD, Ross P, Alm R, Gao N, Kutschke A, Johnstone M, Prince B, Thresher J (2013) Exploring the UDP pocket of LpxC through amino acid analogs. Bioorg Med Chem Lett 23:2362–2367
Barb AW, McClerren AL, Snehelatha K, Reynolds CM, Zhou P, Raetz CR (2007) Inhibition of lipid A biosynthesis as the primary mechanism of CHIR-090 antibiotic activity in Escherichia coli. Biochemistry 46:3793–3802
Lee CJ, Liang X, Chen X, Zeng D, Joo SH, Chung HS, Barb AW, Swanson SM, Nicholas RA, Li Y, Toone EJ (2011) Species-specific and inhibitor-dependent conformations of LpxC: implications for antibiotic design. Chem Biol 18:38–47
Fuhrer T, Heer D, Begemann B, Zamboni N (2011) High-throughput, accurate mass metabolome profiling of cellular extracts by flow injection–time-of-flight mass spectrometry. Anal Chem 83:7074–7080
Patel K, Kumar A, Durani S (2007) Analysis of the structural consensus of the zinc coordination centers of metalloprotein structures. Biochim Biophys Acta Proteins Proteomics 1774:1247–1253
Acknowledgements
Authors are highly grateful to the Pakistan-United States Science and Technology Cooperation Program for granting the financial assistance.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
12539_2018_299_MOESM1_ESM.docx
Table Sl-1 DrugBank features of essential, druggable targets and their predicted sub cellular localizations (DOCX 11 KB)
12539_2018_299_MOESM2_ESM.pdf
Table SI-2 Chemical structure, GOLD scores and AutoDock Vina acquired binding affinities (kJ mol−1) of 249 docked compounds (PDF 733 KB)
12539_2018_299_MOESM3_ESM.docx
Table SI-3 Zn coordination dynamics observed in undocked and docked KpLpxC systems over 12 and 30 ns simulations, respectively (DOCX 12 KB)
12539_2018_299_MOESM4_ESM.tif
Fig. SI-1 Distribution of 204 pathogen-specific, essential proteins in 37 unique metabolic pathways of K. pneumoniae HS11286 (TIF 2052 KB)

12539_2018_299_MOESM6_ESM.tif
Fig. SI-3 Multiple sequence alignment of KpLpxC against PaLpxC, YeLpxC and EcLpxC for active site prediction (TIF 4957 KB)
12539_2018_299_MOESM7_ESM.tif
Fig. SI-4 Superimposed RMSD, RMSF and β-Factor graphs of undocked and docked KpLpxC systems after 12 ns simulations (TIF 7102 KB)
Rights and permissions
About this article
Cite this article
Ahmad, S., Navid, A., Akhtar, A.S. et al. Subtractive Genomics, Molecular Docking and Molecular Dynamics Simulation Revealed LpxC as a Potential Drug Target Against Multi-Drug Resistant Klebsiella pneumoniae. Interdiscip Sci Comput Life Sci 11, 508–526 (2019). https://doi.org/10.1007/s12539-018-0299-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12539-018-0299-y