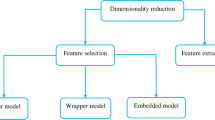
Abstract
XCS is a genetics-based machine learning model that combines reinforcement learning with evolutionary algorithms to evolve a population of classifiers in the form of condition-action rules. Like many other machine learning algorithms, XCS is less effective on high-dimensional data sets. In this paper, we describe a new guided rule discovery mechanisms for XCS, inspired by feature selection techniques commonly used in machine learning. In our approach, feature quality information is used to bias the evolutionary operators. A comprehensive set of experiments is used to investigate how the number of features used to bias the evolutionary operators, population size, and feature ranking technique, affect model performance. Numerical simulations have shown that our guided rule discovery mechanism improves the performance of XCS in terms of accuracy, execution time and more generally in terms of classifier diversity in the population, especially for high-dimensional classification problems. We present a detailed discussion of the effects of model parameters and recommend settings for large scale problems.







Similar content being viewed by others
Notes
GRD-XCS was introduced in [5]. This paper is a revised and a substantially extended version of that paper.
Victorian Partnership for Advanced Computing: http://www.vpac.org.
References
The XCS source code in C is freely available on Illinois Genetic Algorithms Laboratory (IlliGAL) web site: http://illigal.org/category/source-code.
UCI Machine Learning Repository: The Center for Machine Learning and Intelligent Systems at the University of California, Irvine. http://archive.ics.uci.edu/ml
Weka 3, is an open source data mining tool (in java), with a collection of machine learning algorithms developed by Machine Learning Group at University of Waikato. http://www.cs.waikato.ac.nz/ml/weka
Abedini M, Kirley M (2010) A multiple population XCS: evolving condition-action rules based on feature space partitions. In: 2010 IEEE Congress on Evolutionary computation (CEC), pp 1–8, July 2010
Abedini M, Kirley M (2011) Guided rule discovery in XCS for high-dimensional classification problems. In: Proceedings of 24th Australasian artificial intelligence conference. Lecture notes in artificial intelligence, vol 7106
Alon U, Barkai N, Notterman DA, Gishdagger K, Ybarradagger S, Mackdagger D, Levine AJ (1999) Broad patterns of gene expression revealed by clustering analysis of tumor and normal colon tissues probed by oligonucleotide arrays. Proc Natl Acad Sci USA 96:6745–6750
Bacardit J, Krasnogor N (2006) Smart crossover operator with multiple parents for a Pittsburgh learning classifier system. In: Proceedings of the 8th conference on GECCO. ACM, New York, pp 1441–1448
Bacardit J, Stout M, Hirst JD, Sastry K, Llorà à X, Krasnogor N (2007) Automated alphabet reduction method with evolutionary algorithms for protein structure prediction. In: Thierens D, Beyer H-G, Bongard J, Branke J, Clark JA, Cliff D, Congdon CB, Deb K, Doerr B, Kovacs T, Kumar S, Miller JF, Moore J, Neumann F, Pelikan M, Poli R, Sastry K, Stanley KO, Stutzle T, Watson RA, Wegener I (eds) GECCO ’07: Proceedings of the 9th annual conference on Genetic and evolutionary computation, vol 1, London, 7–11 July 2007. ACM Press, New York, pp 346–353
Bacardit J, Stout M, Krasnogor N, Hirst JD, Blazewicz J (2006) Coordination number prediction using learning classifier systems: performance and interpretability. In: Cattolico M (ed) Genetic and evolutionary computation conference, GECCO 2006, proceedings, Seattle, Washington, USA, July 8–12, 2006. ACM, New York, pp 247–254
Bonilla Huerta E, Hernandez Hernandez J, Hernandez Montiel L (2010) A new combined filter-wrapper framework for gene subset selection with specialized genetic operators. In: Advances in pattern recognition. Lecture notes in computer science, vol 6256. Springer, Berlin/Heidelberg, pp 250–259
Butz MV, Goldberg DE, Tharakunnel K (2003) Analysis and improvement of fitness exploitation in XCS: bounding models, tournament selection, and bilateral accuracy. Evol Comput 11:239–277
Butz MV, Pelikan M, Llorà à X, Goldberg DE (2006) Automated global structure extraction for effective local building block processing in XCS. Evol Comput 14:345–380
Butz MV, Wilson SW (2001) An Algorithmic description of XCS. In: Advances in learning classifier systems. Lecture notes in computer science, vol 1996/2001. Springer, Berlin/Heidelberg, pp 267–274
Fernandndez A, Garcianda S, Luengo J, Bernado-Mansilla E, Herrera F (2010) Genetics-based machine learning for rule induction: state of the art, taxonomy, and comparative study. IEEE Trans Evol Comput 14(6):913–941
Golub TR, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, Mesirov JP, Coller H, Loh ML, Downing JR, Caligiuri MA, Bloomfield CD (1999) Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science 286:531–537
Guyon I, Weston J, Barnhill S, Vapnik V (2002) Gene selection for cancer classification using support vector machines. Mach Learn 46:389–422
Hall MA (1998) Correlation-based feature subset selection for machine learning. PhD thesis, University of Waikato, Hamilton, New Zealand
Hedenfalk I, Duggan D, Chen Y, Radmacher M, Bittner M, Simon R, Meltzer P, Gusterson B, Esteller M, Kallioniemi OP, Wilfond B, Borg A, Trent J (2001) Gene-expression profiles in hereditary breast cancer. N Engl J Med 344(8):539–548
Ian EF, Witten H (2005) Data mining: practical machine learning tools and techniques. Morgan Kaufmann series in data management systems, 2 edn. Morgan Kaufmann, Menlo Park
Isabelle Guyon MN, Steve Gunn, Zadeh L (eds) (2006) Feature extraction, foundations and applications. Springer, Berlin
José-Revuelta LMS (2008) A hybrid GA-TS technique with dynamic operators and its application to channel equalization and fiber tracking. In: Jaziri W (ed) Local search techniques: focus on tabu search, I-Tech, Vienna. ISBN 978-3-902613-34-9
Kononenko I (1994) Estimating attributes: analysis and extensions of relief. In: Bergadano F, Raedt LD (eds) Machine learning: Proceedings of the ECML-94, european conference on machine learning, Catania, Italy, April 6–8, 1994. Lecture notes in computer science, vol 784. Springer, Berlin, pp 171–182
Lanzi PL (1997) A study of the generalization capabilities of XCS. In: Bäck T (ed) Proceedings of the 7th international conference on genetic algorithms. Morgan Kaufmann, Menlo Park, pp 418–425
Moore JH, White BC (2006) Exploiting expert knowledge in genetic programming for genome-wide genetic analysis. In: PPSN. Lecture notes in computer science, vol 4193. Springer, Berlin, pp 969–977
Morales-Ortigosa S, Orriols-Puig A, Bernadó-Mansilla E (2008) New crossover operator for evolutionary rule discovery in XCS. In: 8th international conference on hybrid intelligent systems. IEEE Computer Society, pp 867–872
Morales-Ortigosa S, Orriols-Puig A, Bernadó-Mansilla E (2009) Analysis and improvement of the genetic discovery component of XCS. Int Jt Conf Hybrid Intell Syst 6:81–95
Orriols-Puig A, Casillas J, Bernadó-Mansilla E (2008) Genetic-based machine learning systems are competitive for pattern recognition. Evol Intell 1:209–232. doi:10.1007/s12065-008-0013-9
Pang-Ning Tan MSVK (2006) Introduction to data mining. Addison-Wesley Longman Publishing Co., Inc., Chicago
Platt J (1998) Fast training of support vector machines using sequential minimal optimization. In: Schoelkopf B, Burges C, Smola A (eds) Advances in kernel methods—support vector learning. MIT Press, Cambridge
Singh D, Febbo PG, Ross K, Jackson DG, Manola J, Ladd C, Tamayo P, Renshaw AA (2002) Gene expression correlates of clinical prostate cancer behavior. Cancer Cell 1:203–209
Stalph PO, Butz MV, Goldberg DE, Llorà à X (2009) On the scalability of xcs(f). In: Rothlauf F (ed) GECCO. ACM, New York, pp 1315–1322
Sumathi S, Sivanandam SN (2006) Introduction to data mining and its applications. Studies in computational intelligence, vol 29. Springer, Berlin
Wang P, Weise T, Chiong R (2011) Novel evolutionary algorithms for supervised classification problems: an experimental study. Evol Intell 4(1):3–16
Wilson SW (1995) Classifier fitness based on accuracy. Evol Comput 3(2):149–175. http://prediction-dynamics.com/
Wilson SW (1999) Get real! XCS with continuous-valued inputs. In: Lanzi PL, Stolzmann W, Wilson SW (eds) Learning classifier systems, from foundations to applications. Lecture notes in computer science, vol 1813. MIT Press, Cambridge, pp 209–222
Wu F-X, Zhang W, Kusalik A (2006) On Determination of minimum sample size for discovery of temporal gene expression patterns. In: First international multi-symposiums on computer and computational sciences, pp 96–103
Zhang Y-Q, Rajapakse JC (eds) (2008) Machine learning in bioinformatics. Wiley book series on bioinformatics: computational techniques and engineering. 1st edn. John Wiley & Sons, New Jersey
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Abedini, M., Kirley, M. An enhanced XCS rule discovery module using feature ranking. Int. J. Mach. Learn. & Cyber. 4, 173–187 (2013). https://doi.org/10.1007/s13042-012-0085-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13042-012-0085-9